3C5D
 
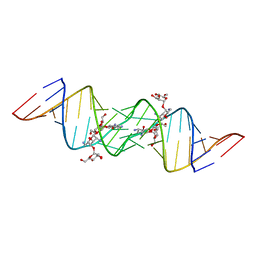 | | Crystal structure of HIV-1 subtype F DIS extended duplex RNA bound to lividomycin | | Descriptor: | 'HIV-1 subtype F genomic RNA, (2R,3S,4S,5S,6R)-2-((2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3S,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R ,5S,6R)-3-AMINO-5-HYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-4-HYDROXY-2-(HYDROXYMET HYL)-TETRAHYDROFURAN-3-YLOXY)-4-HYDROXY-TETRAHYDRO-2H-PYRAN-3-YLOXY)-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-3,4,5-TRIOL, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-01-31 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3C7R
 
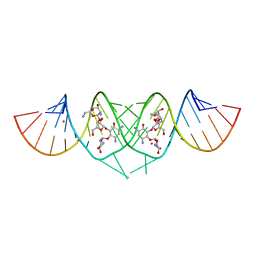 | | Crystal Structure of HIV-1 subtype F DIS extended duplex RNA bound to neomycin | | Descriptor: | HIV-1 subtype F genomic RNA, NEOMYCIN, POTASSIUM ION | | Authors: | Freisz, S, Lang, K, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3DW4
 
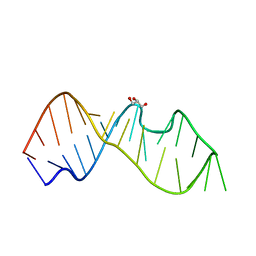 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
4LAR
 
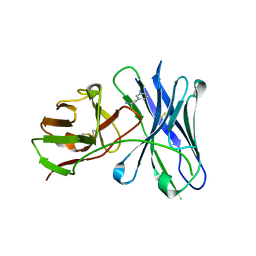 | | Crystal structure of a therapeutic single chain antibody in complex with amphetamine | | Descriptor: | (2S)-1-phenylpropan-2-amine, Single chain antibody fragment scFv6H4 | | Authors: | Celical, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
4LAQ
 
 | | Crystal structure of a therapeutic single chain antibody in the free form | | Descriptor: | D-MALATE, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Celikel, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
3DVZ
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. coli 23 S rRNA | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW7
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
4LAS
 
 | | Crystal structure of a therapeutic single chain antibody in complex with 4-hydroxymethamphetamine | | Descriptor: | 4-[(2S)-2-(methylamino)propyl]phenol, Single chain antibody fragment scFv6H4 | | Authors: | Celical, R, Gokulan, K, Peterson, E.C, Varughese, K.I. | | Deposit date: | 2013-06-20 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural characterization of a therapeutic anti-methamphetamine antibody fragment: oligomerization and binding of active metabolites.
Plos One, 8, 2013
|
|
3DW5
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
7K32
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K33
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
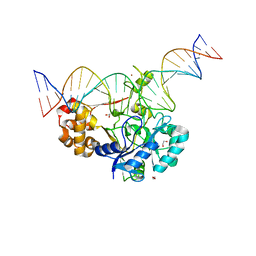 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | Descriptor: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
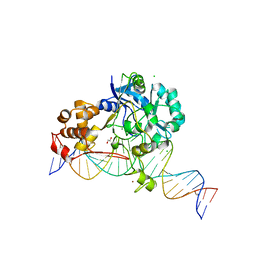 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2WJ8
 
 | | Respiratory Syncitial Virus RiboNucleoProtein | | Descriptor: | BORATE ION, NUCLEOPROTEIN, RNA (5'-R(*CP*CP*CP*CP*CP*C)-3') | | Authors: | Tawar, R.G, Duquerroy, S, Vonrhein, C, Varela, P.F, Damier-Piolle, L, Castagne, N, MacLellan, K, Bedouelle, H, Bricogne, G, Bhella, D, Eleouet, J, Rey, F.A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus
Science, 326, 2009
|
|
4PUT
 
 | |
2VYW
 
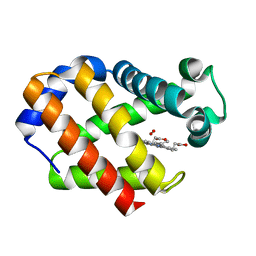 | | Hemoglobin (Hb2) from trematode Fasciola hepatica | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dewilde, S, Ioanitescu, A.I, Kiger, L, Gilany, K, Marden, M.C, Van Doorslaer, S, Vercruysse, J, Pesce, A, Nardini, M, Bolognesi, M, Moens, L. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hemoglobins of the Trematodes Fasciola Hepatica and Paramphistomum Epiclitum: A Molecular Biological, Physico-Chemical, Kinetic, and Vaccination Study.
Protein Sci., 17, 2008
|
|
6E0Y
 
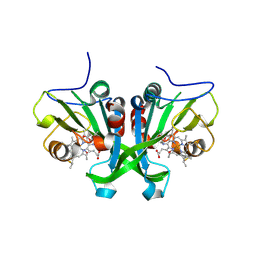 | |
6E0Z
 
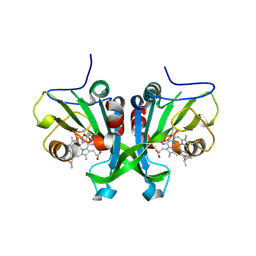 | |
6E17
 
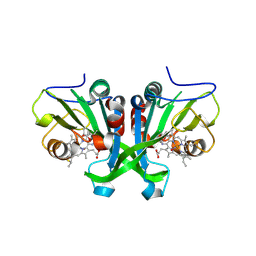 | |
6E0X
 
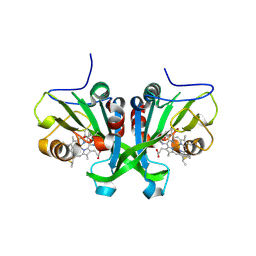 | |
2QZ8
 
 | | Crystal structure of Mycobacterium tuberculosis Leucine response regulatory protein (LrpA) | | Descriptor: | Probable transcriptional regulatory protein | | Authors: | Manchi, C.M.R, Gokulan, K, Ioerger, T, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis LrpA, a leucine-responsive global regulator associated with starvation response.
Protein Sci., 17, 2008
|
|
6AMG
 
 | | cyt P460 of Nitrosomonas sp. AL212 | | Descriptor: | Cytochrome P460, HEME C | | Authors: | Smith, M, Lancaster, K. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Eponymous Cofactors in Cytochrome P460s from Ammonia-Oxidizing Bacteria Are Iron Porphyrinoids Whose Macrocycles Are Dibasic.
Biochemistry, 57, 2018
|
|
1THO
 
 | |
6F5H
 
 | | Crystal structure of USP7 in complex with a 4-hydroxypiperidine based inhibitor | | Descriptor: | 3-[[4-oxidanyl-1-[(3~{R})-3-phenylbutanoyl]piperidin-4-yl]methyl]-6-(2-pyrrolidin-1-ylethylamino)pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, J, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H, Feutron-Burton, S, Rountree, J.S.S. | | Deposit date: | 2017-12-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors.
ACS Med Chem Lett, 9, 2018
|
|
