3E8B
 
 | |
3E89
 
 | |
3E8G
 
 | |
1IIH
 
 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
2RKJ
 
 | | Cocrystal structure of a tyrosyl-tRNA synthetase splicing factor with a group I intron RNA | | Descriptor: | RNA (238-MER), RNA (5'-R(P*GP*CP*UP*U)-3'), Tyrosyl-tRNA synthetase | | Authors: | Paukstelis, P.J, Chen, J.-H, Chase, E, Lambowitz, A.M, Golden, B.L. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA.
Nature, 451, 2008
|
|
2VZ0
 
 | | Pteridine Reductase 1 (PTR1) from Trypanosoma Brucei in complex with NADP and DDD00066641 | | Descriptor: | 6-(4-methylphenyl)quinazoline-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Robinson, D.A, Thompson, S, Sienkiewicz, N, Fairlamb, A.H. | | Deposit date: | 2008-07-29 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development and Validation of a Cytochrome C Coupled Assay for Pteridine Reductase 1 and Dihydrofolate Reductase.
Anal.Biochem., 396, 2010
|
|
2W2S
 
 | | Structure of the Lagos bat virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2WBA
 
 | | Properties of Trypanothione Reductase From T. brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Jones, D, Ariza, A, Chow, W.H.A, Oza, S.L, Fairlamb, A.H. | | Deposit date: | 2009-02-24 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparative Structural, Kinetic and Inhibitor Studies of Trypanosoma Brucei Trypanothione Reductase with T. Cruzi.
Mol.Biochem.Parasitol., 169, 2010
|
|
4TYE
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase | | Descriptor: | Fibroblast growth factor receptor 4, PHOSPHATE ION | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
3E8H
 
 | |
4TYI
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Fibroblast growth factor receptor 4 | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
3SQX
 
 | |
2WOI
 
 | | Trypanothione reductase from Trypanosoma brucei | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WP6
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00071494) | | Descriptor: | (4S)-3-BENZYL-6-CHLORO-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPC
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073357) | | Descriptor: | (4S)-6-CHLORO-3-{2-[4-(FURAN-2-YLCARBONYL)PIPERAZIN-1-YL]ETHYL}-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-05 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
3XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, sorbitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
2WOV
 
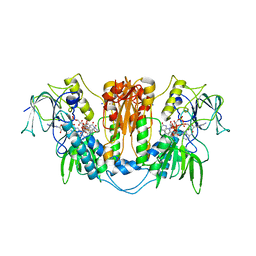 | | Trypanosoma brucei trypanothione reductase with bound NADP. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPE
 
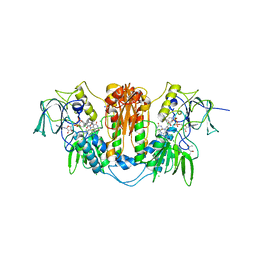 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073359) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPF
 
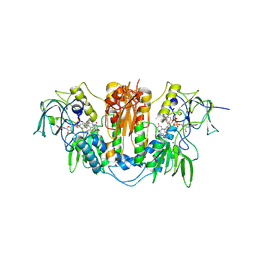 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00085762) | | Descriptor: | 3-[(4S)-6-CHLORO-2-METHYL-4-(4-METHYLPHENYL)QUINAZOLIN-3(4H)-YL]-N,N-DIMETHYLPROPAN-1-AMINE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WSA
 
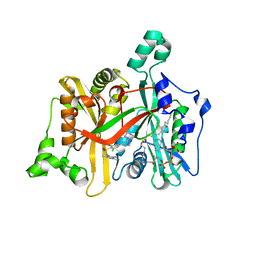 | | Crystal Structure of Leishmania major N-myristoyltransferase (NMT) with bound myristoyl-CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness.
Nature, 464, 2010
|
|
1M3Z
 
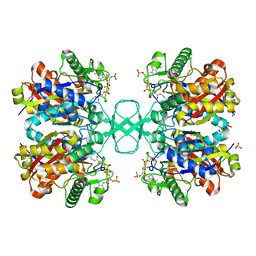 | | Biosynthetic thiolase, C89A mutant, complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
2C21
 
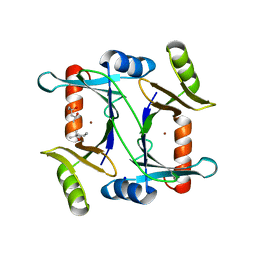 | | Specificity of the Trypanothione-dependednt Leishmania major Glyoxalase I: Structure and biochemical comparison with the human enzyme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, NICKEL (II) ION, ... | | Authors: | Ariza, A, Vickers, T.J, Greig, N, Armour, K.A, Eggleston, I.M, Fairlamb, A.H, Bond, C.S. | | Deposit date: | 2005-09-23 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of the Trypanothione-Dependent Leishmania Major Glyoxalase I: Structure and Biochemical Comparison with the Human Enzyme.
Mol.Microbiol., 59, 2006
|
|
2C07
 
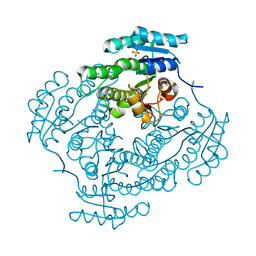 | | Oxoacyl-ACP reductase of Plasmodium falciparum | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER PROTEIN) REDUCTASE, SULFATE ION | | Authors: | Urch, J.E, Wickramasinghe, S.R, Inglis, K.A, Muller, S, Fairlamb, A.H, van Aalten, D.M.F. | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, Inhibition and Structural Studies on 3-Oxoacyl-Acp Reductase from Plasmodium Falciparum, a Key Enzyme in Fatty Acid Biosynthesis.
Biochem.J., 393, 2006
|
|
8FX1
 
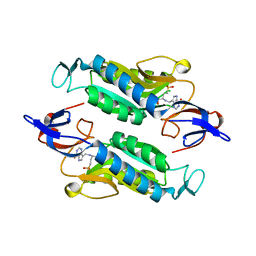 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (R)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWY
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to the dead-end complex xanthine and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, XANTHINE | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
