5Z69
 
 | |
5ZBN
 
 | |
5Z67
 
 | |
4EFO
 
 | | Crystal structure of the ubiquitin-like domain of human TBK1 | | Descriptor: | Serine/threonine-protein kinase TBK1 | | Authors: | Li, J, Li, J, Miyahira, A, Sun, J, Liu, Y, Cheng, G, Liang, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Crystal structure of the ubiquitin-like domain of human TBK1.
Protein Cell, 3, 2012
|
|
7YOX
 
 | |
4ENF
 
 | | Crystal structure of the cap-binding domain of polymerase basic protein 2 from influenza virus A/Puerto Rico/8/34(h1n1) | | Descriptor: | 1,4-BUTANEDIOL, NITRATE ION, Polymerase basic protein 2 | | Authors: | Meng, G, Liu, Y, Zheng, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus
J.Biol.Chem., 288, 2013
|
|
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
4O7X
 
 | |
7WI7
 
 | | Crystal structure of human MCM8/9 complex | | Descriptor: | DNA helicase MCM8, DNA helicase MCM9, ZINC ION | | Authors: | Li, J, Liu, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Crystal structure of human MCM8/9 complex
To Be Published
|
|
2LQ9
 
 | |
2J6W
 
 | | R164N mutant of the RUNX1 Runt domain | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Grembecka, J, Zhe, L, Lukasik, S.M, Liu, Y, Bielnicka, I, Bushweller, J.H, Speck, N.A. | | Deposit date: | 2006-10-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mutation in the S-Switch Region of the Runt Domain Alters the Dynamics of an Allosteric Network Responsible for Cbfbeta Regulation.
J.Mol.Biol., 364, 2006
|
|
6B0Y
 
 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2JVC
 
 | |
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UA3
 
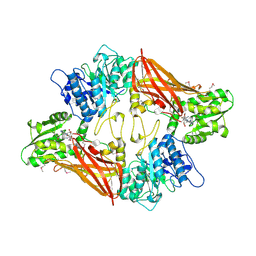 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 in complex with SAH | | Descriptor: | Protein arginine N-methyltransferase 5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-20 | | Release date: | 2011-12-14 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MLC
 
 | | PHD6 domain of MLL3 in complex with histone H4 | | Descriptor: | GLYCEROL, Histone H4, Histone-lysine N-methyltransferase 2C, ... | | Authors: | Dong, A, Liu, Y, Qin, S, Lei, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4.
Nat Commun, 10, 2019
|
|
7FBT
 
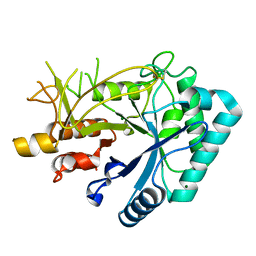 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | Descriptor: | Chitinase, MAGNESIUM ION | | Authors: | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
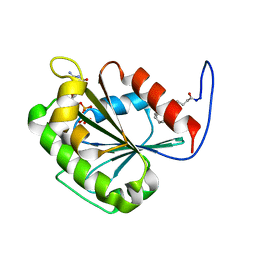
2K5U
 
 | |
5ZUE
 
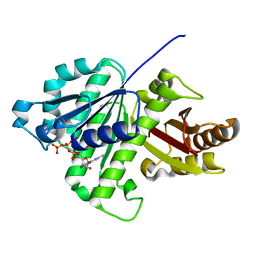 | | GTP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Guan, F, Yu, J, Yu, J, Liu, Y, Li, Y, Feng, X.H, Huang, K.C, Chang, Z, Ye, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division
Elife, 7, 2018
|
|
3UA4
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 5 | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
2OIG
 
 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | Descriptor: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OIE
 
 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
7C3V
 
 | | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dai, W, Ni, Y, Xu, G, Liu, Y, Wang, Y, Zhou, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20042944 Å) | | Cite: | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH)
To Be Published
|
|
6M4Q
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
