8DMF
 
 | | Cryo-EM structure of the ribosome-bound Bacteroides thetaiotaomicron EF-G2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tetracycline resistance protein TetQ | | Authors: | Wang, C, Han, W, Groisman, E.A, Liu, J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.
Embo J., 2022
|
|
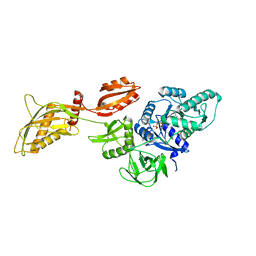
5T0A
 
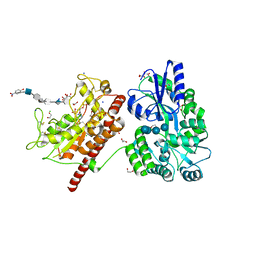 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T05
 
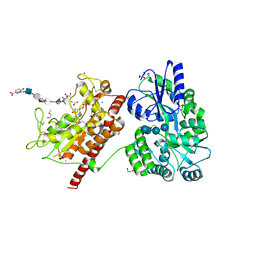 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and IdoA2S containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
3BD9
 
 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
3BEY
 
 | | Crystal structure of the protein O27018 from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium target TT217 | | Descriptor: | Conserved protein O27018 | | Authors: | Kuzin, A.P, Gu, J, Xu, X, Neely, H, Forouhar, F, Owens, L, Mao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the protein O27018 from Methanobacterium thermoautotrophicum.
To be Published
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3BB6
 
 | | Crystal structure of the P64488 protein from E.coli (strain K12). Northeast Structural Genomics Consortium target ER596 | | Descriptor: | Uncharacterized protein yeaR, ZINC ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray structure of the P64488 from E.coli (strain K12).
To be Published
|
|
3BDR
 
 | | Crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus. Northeast Structural Genomics Consortium target TeR13. | | Descriptor: | PHOSPHATE ION, Ycf58 protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus.
To be Published
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMD
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
5TSA
 
 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Zn2+ | | Descriptor: | CADMIUM ION, Membrane protein, ZINC ION | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
3BDU
 
 | | Crystal structure of protein Q6D8G1 at the resolution 1.9 A. Northeast Structural Genomics Consortium target EwR22A. | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein Q6D8G1 at the resolution 1.9 A.
To be Published
|
|
7NTH
 
 | | Structure of TAK1 in complex with compound 54 | | Descriptor: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTI
 
 | | Structure of TAK1 in complex with compound 22 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
3AMP
 
 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMQ
 
 | | E134C-Cellobiose co-crystal of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMS
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
5K0K
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
