7E7C
 
 | |
5HJC
 
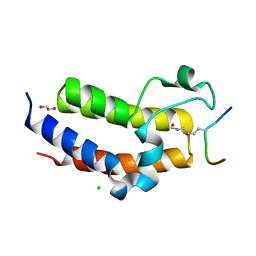 | | BRD3 second bromodomain in complex with histone H3 acetylation at K18 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, CHLORIDE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
1BM6
 
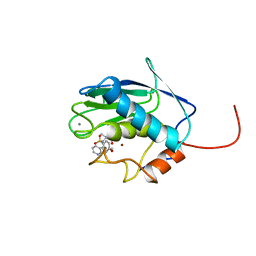 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Li, Y, Zhang, X, Melton, R, Ganu, V, Gonnella, N.C. | | Deposit date: | 1998-07-29 | | Release date: | 1999-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin-1 complexed to a potent, nonpeptidic inhibitor.
Biochemistry, 37, 1998
|
|
3FF7
 
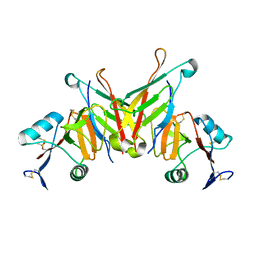 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | ACETIC ACID, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
6U18
 
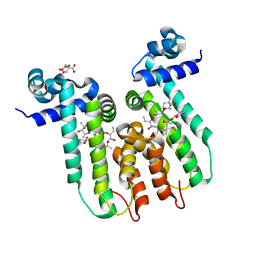 | | Directed evolution of a biosensor selective for the macrolide antibiotic clarithromycin | | Descriptor: | CITRATE ANION, CLARITHROMYCIN, Erythromycin resistance repressor protein | | Authors: | Li, Y, Reed, M, Wright, H.T, Cropp, T.A, Williams, G. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Genetically Encoded Biosensors for Reporting the Methyltransferase-Dependent Biosynthesis of Semisynthetic Macrolide Antibiotics.
Acs Synth Biol, 2021
|
|
2KTQ
 
 | | OPEN TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Li, Y, Waksman, G. | | Deposit date: | 1998-07-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation.
EMBO J., 17, 1998
|
|
3FF9
 
 | | Structure of NK cell receptor KLRG1 | | Descriptor: | Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
3KTQ
 
 | |
2L92
 
 | |
8W8F
 
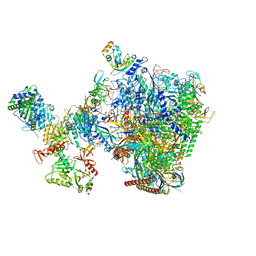 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8EFN
 
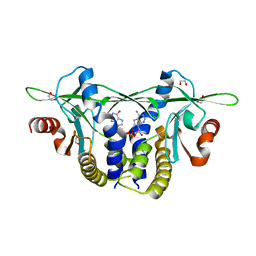 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8W8E
 
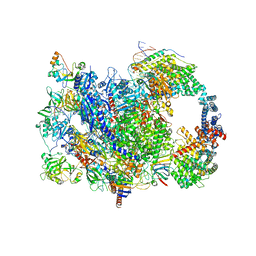 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
3L6F
 
 | |
2QM4
 
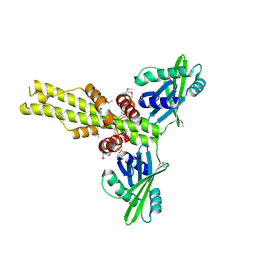 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
5HJD
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K18 | | Descriptor: | COPPER (II) ION, Protein AF-9, SULFATE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain.
Mol.Cell, 62, 2016
|
|
5VZT
 
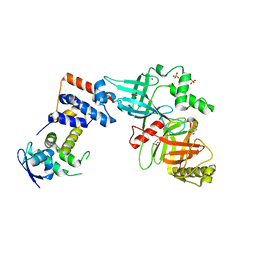 | | Crystal structure of the Skp1-FBXO31 complex | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VZU
 
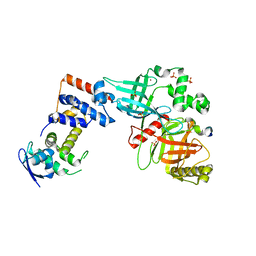 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2NB8
 
 | |
2HB0
 
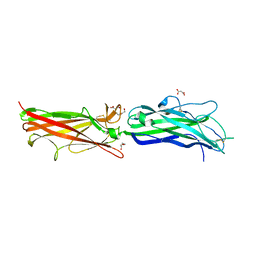 | | Crystal Structure of CfaE, the Adhesive Subunit of CFA/I Fimbria of Enterotoxigenic Escherichia coli | | Descriptor: | CFA/I fimbrial subunit E, DI(HYDROXYETHYL)ETHER, MALONATE ION | | Authors: | Li, Y.F, Xia, D, Poole, S, Rasulova, F, Savarino, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
8GJY
 
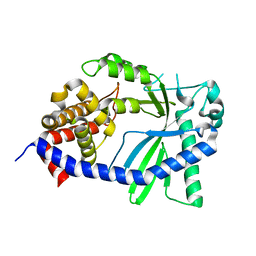 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7E7A
 
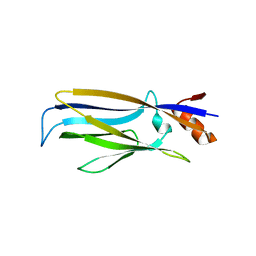 | | Crystal structure of apo ENL YEATS domain T3 mutant | | Descriptor: | Protein ENL | | Authors: | Li, Y, Li, H. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of ENL YEATS domain T1 mutant in complex with histone H3 acetylation at K27
To Be Published
|
|
6M62
 
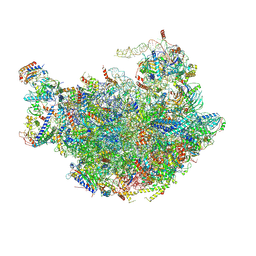 | |
5YYF
 
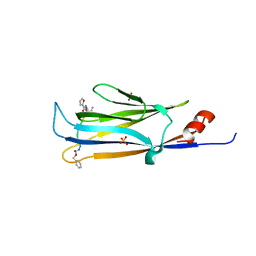 | |
1VF6
 
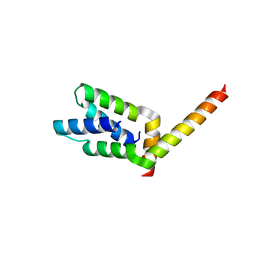 | | 2.1 Angstrom crystal structure of the PALS-1-L27N and PATJ L27 heterodimer complex | | Descriptor: | MAGUK p55 subfamily member 5, PALS1-associated tight junction protein | | Authors: | Li, Y, Lavie, A, Margolis, B, Karnak, D. | | Deposit date: | 2004-04-09 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for L27 domain-mediated assembly of signaling and cell polarity complexes.
Embo J., 23, 2004
|
|
2LS1
 
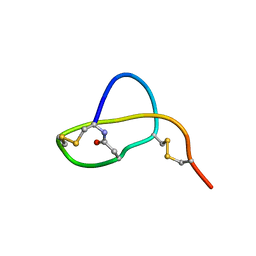 | | Structure of Sviceucin, an antibacterial type I lasso peptide from Streptomyces sviceus | | Descriptor: | Uncharacterized protein | | Authors: | Li, Y, Ducasse, R, Blond, A, Zirah, S, Goulard, C, Lescop, E, Guittet, E, Pernodet, J, Rebuffat, S. | | Deposit date: | 2012-04-18 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure and biosynthesis of Sviceucin, an antibacterial type I lasso peptide from Streptomyces sviceus
To be Published
|
|
