2AYD
 
 | | Crystal Structure of the C-terminal WRKY domainof AtWRKY1, an SA-induced and partially NPR1-dependent transcription factor | | Descriptor: | SUCCINIC ACID, WRKY transcription factor 1, ZINC ION | | Authors: | Duan, M.R, Nan, J, Li, Y, Su, X.D. | | Deposit date: | 2005-09-07 | | Release date: | 2006-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein.
Nucleic Acids Res., 35, 2007
|
|
1Y00
 
 | | Solution structure of the Carbon Storage Regulator protein CsrA | | Descriptor: | Carbon storage regulator | | Authors: | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-13 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
2X9L
 
 | | Crystal structure of deacetylase-bog complex in biosynthesis pathway of teicoplanin. | | Descriptor: | N-ACYL GLM PEUDO-TEICOPLANIN DEACETYLASE, ZINC ION, octyl beta-D-glucopyranoside | | Authors: | Chan, H.C, Huang, Y.T, Lyu, S.Y, Huang, C.J, Li, Y.S, Liu, Y.C, Chou, C.C, Tsai, M.D, Li, T.L. | | Deposit date: | 2010-03-23 | | Release date: | 2011-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Regioselective Deacetylation Based on Teicoplanin-Complexed Orf2 Crystal Structures.
Mol.Biosyst., 7, 2011
|
|
1XC5
 
 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X9S
 
 | | T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-CTP as the incoming nucleotide. | | Descriptor: | 5'-D(*CP*CP*CP*(AFG)P*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*T*GP*GP*TP*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1N3B
 
 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
1XK6
 
 | | Crystal Structure- P1 form- of Escherichia coli Crotonobetainyl-CoA: carnitine CoA Transferase (CaiB) | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
2WDW
 
 | | The Native Crystal Structure of the Primary Hexose Oxidase (Dbv29) in Antibiotic A40926 Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PUTATIVE HEXOSE OXIDASE | | Authors: | Liu, Y.-C, Li, Y.-S, Lyu, S.-Y, Chen, Y.-H, Chan, H.-C, Huang, C.-J, Huang, Y.-T, Chen, G.-H, Chou, C.-C, Tsai, M.-D, Li, T.-L. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Interception of Teicoplanin Oxidation Intermediates Yields New Antimicrobial Scaffolds
Nat.Chem.Biol., 7, 2011
|
|
1YNI
 
 | | Crystal Structure of N-Succinylarginine Dihydrolase, AstB, bound to Substrate and Product, an Enzyme from the Arginine Catabolic Pathway of Escherichia coli | | Descriptor: | N~2~-(3-CARBOXYPROPANOYL)-L-ARGININE, POTASSIUM ION, Succinylarginine Dihydrolase | | Authors: | Tocilj, A, Schrag, J.D, Li, Y, Schneider, B.L, Reitzer, L, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-succinylarginine dihydrolase AstB, bound to substrate and product, an enzyme from the arginine catabolic pathway of Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
9AVA
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
6BC8
 
 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | Descriptor: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5JUW
 
 | | complex of Dot1l with SS148 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Li, Y, Spurr, S.S, Bayle, E.D, Fish, P.V, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complex of Dot1l with SS148
To Be Published
|
|
6BI7
 
 | | Crystal structure of Rev7-WT/Rev3 as a monomer under high-salt conditions | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Rizzo, A.A, Korzhnev, D.M, Hao, B, Li, Y. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4G0U
 
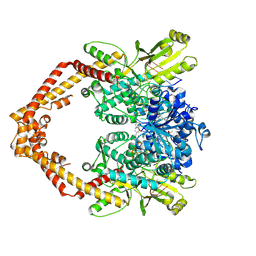 | | Human topoisomerase IIbeta in complex with DNA and amsacrine | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
5KO9
 
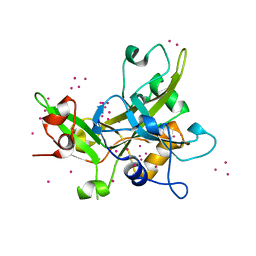 | | Crystal Structure of the SRAP Domain of Human HMCES Protein | | Descriptor: | Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2XAD
 
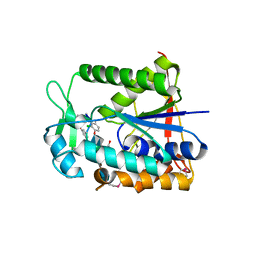 | | Crystal structure of deacetylase-teicoplanin complex in biosynthesis pathway of teicoplanin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Chan, H.C, Huang, Y.T, Lyu, S.Y, Huang, C.J, Li, Y.S, Liu, Y.C, Chou, C.C, Tsai, M.D, Li, T.L. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Regioselective Deacetylation Based on Teicoplanin-Complexed Orf2 Crystal Structures.
Mol.Biosyst., 7, 2011
|
|
7R5F
 
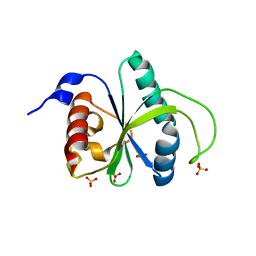 | | Crystal structure of YTHDF2 with compound YLI_DF_012 | | Descriptor: | 5-azanyl-6-methyl-1~{H}-pyrimidine-2,4-dione, CHLORIDE ION, SULFATE ION, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7VCT
 
 | | Human p97 single hexamer conformer III with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7R5W
 
 | | Crystal structure of YTHDF2 with compound YLI_DF_029 | | Descriptor: | 6-cyclopropyl-1H-pyrimidine-2,4-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
4G0V
 
 | | Human topoisomerase iibeta in complex with DNA and mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
1T0F
 
 | | Crystal Structure of the TnsA/TnsC(504-555) complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MALONIC ACID, ... | | Authors: | Ronning, D.R, Li, Y, Perez, Z.N, Ross, P.D, Hickman, A.B, Craig, N.L, Dyda, F. | | Deposit date: | 2004-04-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Embo J., 23, 2004
|
|
5X29
 
 | |
4G0W
 
 | | Human topoisomerase iibeta in complex with DNA and ametantrone | | Descriptor: | 1,4-bis({2-[(2-hydroxyethyl)amino]ethyl}amino)anthracene-9,10-dione, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
1IJI
 
 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
2KFV
 
 | | Structure of the amino-terminal domain of human FK506-binding protein 3 / Northeast Structural Genomics Consortium Target HT99A | | Descriptor: | FK506-binding protein 3 | | Authors: | Sunnerhagen, M, Davis, T, Gutmanas, A, Fares, C, Ouyang, H, Lemak, A, Li, Y, Weigelt, J, Bountra, C, Edwards, A, Arrowsmith, C.H, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of FK506-binding protein 3
To be Published
|
|
