7Y72
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface) | | Descriptor: | Fab E7 heavy chain, Fab E7 light chain, Spike glycoprotein | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
7YEQ
 
 | |
7Y71
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
1APM
 
 | | 2.0 ANGSTROM REFINED CRYSTAL STRUCTURE OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE COMPLEXED WITH A PEPTIDE INHIBITOR AND DETERGENT | | Descriptor: | N-OCTANE, PEPTIDE INHIBITOR PKI(5-24), cAMP-DEPENDENT PROTEIN KINASE | | Authors: | Knighton, D.R, Bell, S.M, Zheng, J, Teneyck, L.F, Xuong, N.-H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1993-01-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with a peptide inhibitor and detergent.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
5Z7G
 
 | | Crystal structure of TAX1BP1 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Tax1-binding protein 1 | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q, Wang, Y.L, Hu, S.C. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZKT
 
 | |
1BTM
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Delboni, L.F, Mande, S.C, Hol, W.G.J. | | Deposit date: | 1995-11-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci., 4, 1995
|
|
1BL4
 
 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BA7
 
 | | SOYBEAN TRYPSIN INHIBITOR | | Descriptor: | TRYPSIN INHIBITOR (KUNITZ) | | Authors: | De Meester, P, Brick, P, Lloyd, L.F, Blow, D.M, Onesti, S. | | Deposit date: | 1998-04-22 | | Release date: | 1998-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Kunitz-type soybean trypsin inhibitor (STI): implication for the interactions between members of the STI family and tissue-plasminogen activator.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1B4M
 
 | | NMR STRUCTURE OF APO CELLULAR RETINOL-BINDING PROTEIN II, 24 STRUCTURES | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II | | Authors: | Lu, J, Lin, C.-L, Tang, C, Ponder, J.W, Kao, J.L.F, Cistola, D.P, Li, E. | | Deposit date: | 1998-12-23 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure and dynamics of rat apo-cellular retinol-binding protein II in solution: comparison with the X-ray structure.
J.Mol.Biol., 286, 1999
|
|
7YX3
 
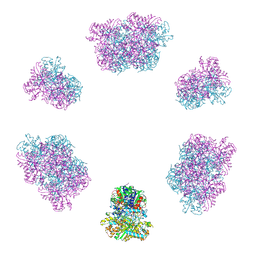 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
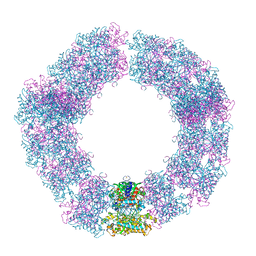 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX5
 
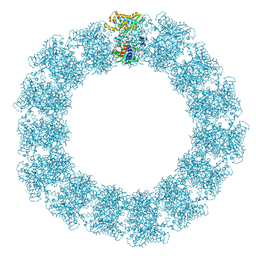 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
1C87
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1C72
 
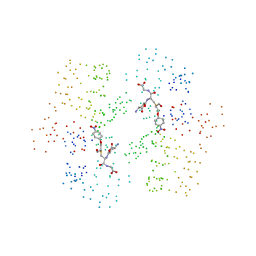 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
1C85
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO)-BENZOIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-BENZOIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1C88
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1C83
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 6-(OXALYL-AMINO)-1H-INDOLE-5-CARBOXYLIC ACID | | Descriptor: | 6-(OXALYL-AMINO)-1H-INDOLE-5-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P. | | Deposit date: | 2000-04-14 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1C84
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH 3-(OXALYL-AMINO)-NAPHTHALENE-2-CARBOXLIC ACID | | Descriptor: | 3-(OXALYL-AMINO)-NAPHTHALENE-2-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Andersen, H.S, Iversen, L.F, Branner, S, Rasmussen, H.B, Moller, N.P. | | Deposit date: | 2000-04-14 | | Release date: | 2000-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2-(oxalylamino)-benzoic acid is a general, competitive inhibitor of protein-tyrosine phosphatases.
J.Biol.Chem., 275, 2000
|
|
1C86
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B (R47V,D48N) COMPLEXED WITH 2-(OXALYL-AMINO-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID | | Descriptor: | 2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE PHOSPHATASE 1B) | | Authors: | Iversen, L.F, Andersen, H.S, Mortensen, S.B, Moller, N.P. | | Deposit date: | 2000-04-16 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of a low molecular weight, nonphosphorus, nonpeptide, and highly selective inhibitor of protein-tyrosine phosphatase 1B.
J.Biol.Chem., 275, 2000
|
|
1BX6
 
 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1BKX
 
 | | A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY | | Descriptor: | ADENOSINE MONOPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Cox, S, Xuong, N, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility.
Structure, 5, 1997
|
|
4EGS
 
 | |
4GBY
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | Descriptor: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | Descriptor: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
