1MBC
 
 | |
2MA2
 
 | | Solution structure of RasGRP2 EF hands bound to calcium | | Descriptor: | RAS guanyl-releasing protein 2 | | Authors: | Kuriyan, J, Iwig, J, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J, Wemmer, D, Roose, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
2TPR
 
 | |
1A06
 
 | | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE | | Authors: | Kuriyan, J, Goldberg, J. | | Deposit date: | 1997-12-09 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I.
Cell(Cambridge,Mass.), 84, 1996
|
|
3KK9
 
 | | CaMKII Substrate Complex B | | Descriptor: | Calcium/calmodulin dependent protein kinase II | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-04 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KK8
 
 | | CaMKII Substrate Complex A | | Descriptor: | Calcium/calmodulin dependent protein kinase II, MAGNESIUM ION | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-04 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KL8
 
 | | CaMKIINtide Inhibitor Complex | | Descriptor: | Calcium/calmodulin dependent protein kinase II, Calcium/calmodulin-dependent protein kinase II inhibitor 1 | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.372 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7WJ3
 
 | | Crystal structure of HLA-C*1402 complexed with 4-mer lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, 4-mer lipopeptide, Beta-2-microglobulin, ... | | Authors: | Kuroha, J, Morita, D, Asa, M, Sugita, M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56343615 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
7WJ2
 
 | | Crystal structure of HLA-C*1402 complexed with 8-mer HIV gag peptide | | Descriptor: | 1,2-ETHANEDIOL, 8-mer peptide, ACETATE ION, ... | | Authors: | Kuroha, J, Morita, D, Asa, M, Sugita, M. | | Deposit date: | 2022-01-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of N-myristoylated lipopeptide-bound HLA class I complexes indicate reorganization of B-pocket architecture upon ligand binding.
J.Biol.Chem., 298, 2022
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
1J3C
 
 | |
1J3X
 
 | |
1J3D
 
 | |
1TRB
 
 | |
5Y95
 
 | | Haddock model of mSIN3B PAH1 domain | | Descriptor: | Paired amphipathic helix protein Sin3b, propan-2-yl (3R,6S,9aS)-3-ethyl-8-(3-methylbutyl)-6-(2-methylsulfanylethyl)-4,7-bis(oxidanylidene)-9,9a-dihydro-6H-pyrazino[2,1-c][1,2,4]oxadiazine-1-carboxylate | | Authors: | Kurita, J, Hirao, Y, Nishimura, Y. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A mimetic of the mSin3-binding helix of NRSF/REST ameliorates abnormal pain behavior in chronic pain models.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4GS3
 
 | | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tencongensis solved ab initio | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Liebschner, D, Brzezinski, K, Dauter, M, Dauter, Z, Nowak, M, Kur, J, Olszewski, M. | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Dimeric structure of the N-terminal domain of PriB protein from Thermoanaerobacter tengcongensis solved ab initio.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3HP4
 
 | | Crystal structure of psychrotrophic esterase EstA from Pseudoalteromonas sp. 643A inhibited by monoethylphosphonate | | Descriptor: | GDSL-esterase | | Authors: | Brzuszkiewicz, A, Nowak, E, Dauter, Z, Dauter, M, Cieslinski, H, Kur, J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of EstA esterase from psychrotrophic Pseudoalteromonas sp. 643A covalently inhibited by monoethylphosphonate.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2AXW
 
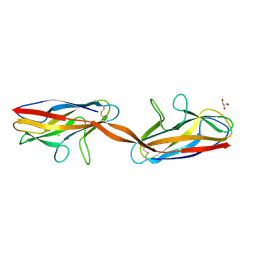 | | Structure of DraD invasin from uropathogenic Escherichia coli | | Descriptor: | CHLORIDE ION, DraD invasin, GLYCEROL | | Authors: | Jedrzejczak, R, Dauter, Z, Dauter, M, Piatek, R, Zalewska, B, Mroz, M, Bury, K, Nowicki, B, Kur, J. | | Deposit date: | 2005-09-06 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped beta-tails.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1EH4
 
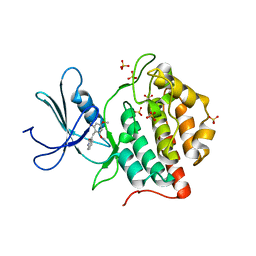 | | BINARY COMPLEX OF CASEIN KINASE-1 FROM S. POMBE WITH AN ATP COMPETITIVE INHIBITOR, IC261 | | Descriptor: | 3-[(2,4,6-TRIMETHOXY-PHENYL)-METHYLENE]-INDOLIN-2-ONE, CASEIN KINASE-1, SULFATE ION | | Authors: | Mashhoon, N, Demaggio, A.J, Tereshko, V, Bergmeier, S.C, Egli, M, Hoekstra, M.F, Kuret, J. | | Deposit date: | 2000-02-18 | | Release date: | 2001-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Conformation-Selective Casein Kinase-1 Inhibitor
J.Biol.Chem., 275, 2000
|
|
7SA7
 
 | |
1EQN
 
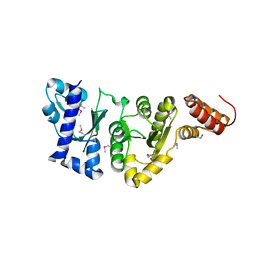 | | E.COLI PRIMASE CATALYTIC CORE | | Descriptor: | DNA PRIMASE | | Authors: | Podobnik, M, McInerney, P, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-04-05 | | Release date: | 2000-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases.
J.Mol.Biol., 300, 2000
|
|
2GS6
 
 | | Crystal Structure of the active EGFR kinase domain in complex with an ATP analog-peptide conjugate | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, Peptide, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2RF9
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
5WDO
 
 | | H-Ras bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
