8UF7
 
 | | Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin | | Descriptor: | POmAb Heavy Chain, POmAb Light Chain, Prothrombin | | Authors: | Kumar, S, Summers, B, Basore, K, Pozzi, N. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and functional basis of prothrombin recognition by a type I antiprothrombin antiphospholipid antibody.
Blood, 143, 2024
|
|
6CA8
 
 | |
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
8X1H
 
 | |
6A8D
 
 | | Crystal Structure of Chlamydomonas reinhardtii ARF | | Descriptor: | ARF/SAR superfamily small monomeric GTP binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kumari, S, Goel, M, Kateriya, S, Sharma, P. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of Chlamydomonas reinhardtii Arf
To Be Published
|
|
6A88
 
 | | Crystal Structure of T. gondii prolyl tRNA synthetase with Febrifugine and ATP Analog | | Descriptor: | 3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kumari, S, Mishra, S, Yogavel, M, Sharma, A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Conformational heterogeneity in apo and drug-bound structures of Toxoplasma gondii prolyl-tRNA synthetase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JP9
 
 | | Crsytal structure of a XMP complexed ATPPase subunit of M. jannaschii GMP synthetase | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B, MALONIC ACID, TRIETHYLENE GLYCOL, ... | | Authors: | Shivakumarasamy, S, Balaram, H. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic Insights into the Functioning of a Two-Subunit GMP Synthetase, an Allosterically Regulated, Ammonia Channeling Enzyme.
Biochemistry, 61, 2022
|
|
2F8Q
 
 | | An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 | | Descriptor: | MAGNESIUM ION, alkaline thermostable endoxylanase | | Authors: | Ramakumar, S, Manikandan, K, Bhardwaj, A, Ghosh, A, Reddy, V.S. | | Deposit date: | 2005-12-03 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions.
Protein Sci., 15, 2006
|
|
2FGL
 
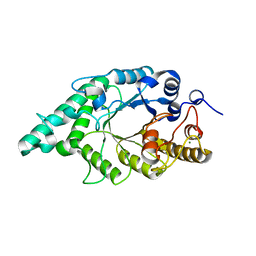 | | An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 | | Descriptor: | MAGNESIUM ION, alkaline thermostable endoxylanase, alpha-D-xylopyranose, ... | | Authors: | Ramakumar, S, Manikandan, K, Bhardwaj, A, Reddy, V.S, Lokanath, N.K, Ghosh, A. | | Deposit date: | 2005-12-22 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of native and xylosaccharide-bound alkali thermostable xylanase from an alkalophilic Bacillus sp. NG-27: structural insights into alkalophilicity and implications for adaptation to polyextreme conditions.
Protein Sci., 15, 2006
|
|
5XLU
 
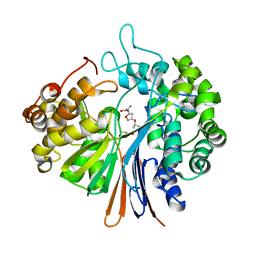 | | High Resolution Crystal Structure of Bacillus Licheniformis Gamma Glutamyl Transpeptidase with Acivicin | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumari, S, Goel, M, Gupta, R, Pal, R. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High Resolution Crystal Structure of Bacillus Licheniformis Gamma Glutamyl Transpeptidase with Acivicin
To Be Published
|
|
5Y8X
 
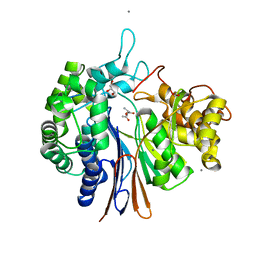 | | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kumari, S, Goel, M, Pal, R, Gupta, R. | | Deposit date: | 2017-08-21 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Bacillus licheniformis Gamma glutamyl transpeptidase with Azaserine
To Be Published
|
|
5ZDI
 
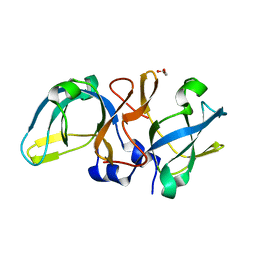 | |
8I7E
 
 | | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Kumar, N, Dilawari, R, Chaubey, G.K, Modanwal, R, Talukdar, S, Dhiman, A, Chaudhary, S, Patidar, A, Kumar, A, Raje, C.I, Raje, M, Kumaran, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A
To Be Published
|
|
3E9K
 
 | | Crystal structure of Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex | | Descriptor: | 3-Hydroxyhippuric acid, Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Kumar, S, Gawandi, V, Momany, C, Phillips, R.S. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity.
J.Med.Chem., 52, 2009
|
|
4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7X7N
 
 | | 3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, Synthetic peptide SIH-5 | | Authors: | Khatri, B, Pramanick, I, Malladi, S.K, Rajmani, R.S, Kumar, S, Ghosh, P, Sengupta, N, Rahisuddin, R, Kumaran, S, Ringe, R.P, Varadarajan, R, Dutta, S, Chatterjee, J. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | A dimeric proteomimetic prevents SARS-CoV-2 infection by dimerizing the spike protein.
Nat.Chem.Biol., 18, 2022
|
|
1BJF
 
 | |
1UBQ
 
 | |
1HIG
 
 | | THREE-DIMENSIONAL STRUCTURE OF RECOMBINANT HUMAN INTERFERON-GAMMA. | | Descriptor: | INTERFERON-GAMMA | | Authors: | Ealick, S.E, Cook, W.J, Vijay-Kumar, S, Carson, M, Nagabhushan, T.L, Trotta, P.P, Bugg, C.E. | | Deposit date: | 1991-10-03 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of recombinant human interferon-gamma.
Science, 252, 1991
|
|
3BM5
 
 | | Crystal structure of O-acetyl-serine sulfhydrylase from Entamoeba histolytica in complex with cysteine | | Descriptor: | CYSTEINE, Cysteine synthase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Krishna, C, Kumar, M, Kumar, S, Gourinath, S. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of native O-acetyl-serine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: structural evidence for cysteine binding and lack of interactions with serine acetyl transferase.
Proteins, 72, 2008
|
|
3OIH
 
 | | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, M, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the complex of xylanase-alpha-amylase inhibitor Protein (XAIP-I) with trehalose at 1.87 A resolution
To be Published
|
|
3FG5
 
 | | Crystal structure determination of a ternary complex of phospholipase A2 with a pentapeptide FLSYK and Ajmaline at 2.5 A resolution | | Descriptor: | AJMALINE, Group II Phospholipase A2, pentapeptide FLSYK | | Authors: | Kumar, M, Kumar, S, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure determination of a ternary complex of phospholipase A2 with a pentapeptide FLSYK and Ajmaline at 2.5 A resolution
To be Published
|
|
3S8H
 
 | | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution | | Descriptor: | 3-HYDROXY-PROPANOIC ACID, Dihydrodipicolinate synthase | | Authors: | Kumar, M, Kaur, N, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-28 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of dihydrodipicolinate synthase complexed with 3-Hydroxypropanoic acid(HPA)at 2.70 A resolution
TO BE PUBLISHED
|
|
3PUO
 
 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
8IYO
 
 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
