6F58
 
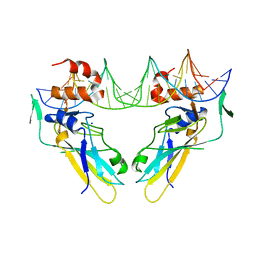 | | Crystal structure of human Brachyury (T) in complex with DNA | | Descriptor: | Brachyury protein, DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*TP*T)-3'), SODIUM ION | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of human Brachyury (T) in complex with DNA
To Be Published
|
|
5LBA
 
 | | Crystal structure of human RECQL5 helicase in complex with DSPL fragment(1-cyclohexyl-3-(oxolan-2-ylmethyl)urea, SGC - Diamond XChem I04-1 fragment screening. | | Descriptor: | 1-cyclohexyl-3-[[(2~{R})-oxolan-2-yl]methyl]urea, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, ... | | Authors: | Newman, J.A, Aitkenhead, H, Talon, R, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human RECQL5 helicase in complex with 3D fragment (1-cyclohexyl-3-(oxolan-2-ylmethyl)urea)
To be published
|
|
6F59
 
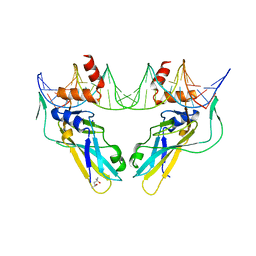 | | Crystal structure of human Brachyury (T) G177D variant in complex with DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Brachyury protein, DNA (26-MER), ... | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Crystal structure of human Brachyury (T) G177D variant in complex with DNA
To Be Published
|
|
7N7Y
 
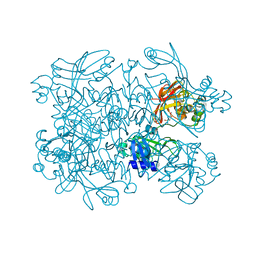 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z18197050 | | Descriptor: | Uridylate-specific endoribonuclease, methyl 4-sulfamoylbenzoate | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7U
 
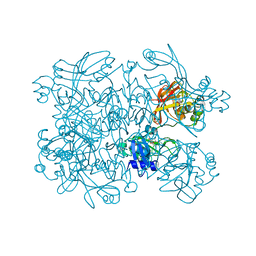 | | Crystal Structure of SARS-CoV-2 NendoU in complex with LIZA-7 | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7W
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with CSC000178569 | | Descriptor: | N-(2-fluorophenyl)-N'-methylurea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7R
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2472938267 | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
4NR2
 
 | | Crystal structure of STK4 (MST1) SARAH domain | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 4 | | Authors: | Chaikuad, A, Krojer, T, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of STK4 (MST1) SARAH domain
To be Published
|
|
7ZMM
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2-001, ZINC ION | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZML
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G1-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G1-001, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMV
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G5-006 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G5-006, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7OCB
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[3-(1,3-dihydroisoindol-2-yl)propoxy]-2N-[2-(dimethylamino)ethyl]-6-methoxy-4N-(1-propan-2-ylpiperidin-4-yl)quinazoline-2,4-diamine, CHLORIDE ION, ... | | Authors: | Johansson, C, Krojer, T, Park, K, Xiong, Y, Jin, J, Oppermann, U. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B
To Be Published
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7OZX
 
 | | Structure of human galactokinase 1 bound with azepan-1-yl(2,6-difluorophenyl)methanone | | Descriptor: | (azepan-1-yl)(2,6-difluorophenyl)methanone, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human galactokinase 1 bound with azepan-1-yl(2,6-difluorophenyl)methanone
To Be Published
|
|
5IGN
 
 | | Crystal structure of human BRD9 bromodomain in complex with LP99 chemical probe | | Descriptor: | Bromodomain-containing protein 9, N-[(2R,3S)-2-(4-chlorophenyl)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxopiperidin-3-yl]-2-methylpropane-1-sulfonamide | | Authors: | Tallant, C, Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-02-28 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with LP99 chemical probe
To Be Published
|
|
4TXC
 
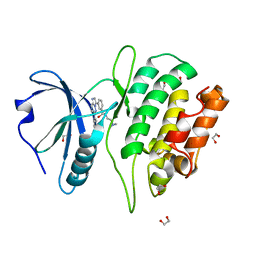 | | Crystal Structure of DAPK1 kinase domain in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-{3-[(R)-{[2-(dimethylamino)ethyl]amino}(hydroxy)methyl]phenyl}imidazo[1,2-b]pyridazin-6-yl)-2-methoxyphenol, Death-associated protein kinase 1 | | Authors: | Sorrell, F.J, Krojer, T, Wilbek, T.S, Skovgaard, T, Berthelsen, J, Dixon-Clarke, S, Chalk, R, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Stromgaard, K, Knapp, S. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Identification and characterization of a small-molecule inhibitor of death-associated protein kinase 1.
Chembiochem, 16, 2015
|
|
4NR7
 
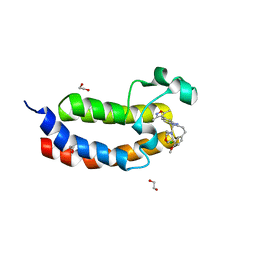 | | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Krojer, T, Nowak, R, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an isoxazolyl-benzimidazole ligand
To be Published
|
|
4GD4
 
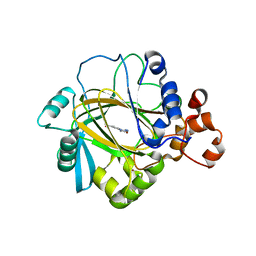 | | Crystal Structure of JMJD2A Complexed with Inhibitor | | Descriptor: | 2-(1H-pyrazol-3-yl)pyridine-4-carboxylic acid, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | Authors: | King, O.N.F, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, McDonough, M.A, Schofield, C.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of JMJD2A Complexed with Inhibitor
TO BE PUBLISHED
|
|
5LF9
 
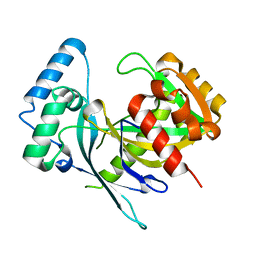 | | Crystal structure of human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22 | | Authors: | Tallant, C, Siejka, P, Mathea, S, Shrestha, L, Krojer, T, Srikannathasan, V, Elkins, J.M, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human NUDT22
To Be Published
|
|
7ZMN
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-048 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-048, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMR
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G2*-011 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G2*-011, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZMO
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G3-052 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G3-052, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
