7UAV
 
 | | Structure of Clostridium botulinum prophage Tad1 in apo state | | Descriptor: | ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
7UAW
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
7UN9
 
 | | SfSTING with c-di-GMP double fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UN8
 
 | | SfSTING with c-di-GMP single fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UNA
 
 | | SfSTING with cGAMP (masked) | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7T27
 
 | | Structure of phage FBB1 anti-CBASS nuclease Acb1-3'3'-cGAMP complex in post reaction state | | Descriptor: | Acb1, SULFATE ION, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-2-[[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphoryl]oxymethyl]-4-oxidanyl-oxolan-3-yl]oxy-sulfanyl-phosphinic acid | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T26
 
 | | Structure of phage FBB1 anti-CBASS nuclease Acb1 in apo state | | Descriptor: | Acb1 | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7T28
 
 | | Structure of phage Bsp38 anti-Pycsar nuclease Apyc1 in apo state | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2R
 
 | | Structure of Paenibacillus sp. J14 Apyc1 | | Descriptor: | Apyc1, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7U2S
 
 | | Structure of Paenibacillus xerothermodurans Apyc1 in the apo state | | Descriptor: | Apyc1, ZINC ION | | Authors: | Hobbs, S.J, Wein, T, Lu, A, Morehouse, B.R, Schnabel, J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-02-24 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phage anti-CBASS and anti-Pycsar nucleases subvert bacterial immunity.
Nature, 605, 2022
|
|
7TQQ
 
 | | Structure of human TREX1-DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*GP*GP*CP*CP*GP*GP*CP*CP*AP*TP*C)-3'), Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
7TQN
 
 | |
7TQO
 
 | |
7TQP
 
 | | Structure of human TREX1 | | Descriptor: | SULFATE ION, Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
7R65
 
 | |
8DP6
 
 | |
8DP7
 
 | |
5UQD
 
 | | DPY-21 in complex with Fe(II) and alpha-Ketoglutarate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-OXOGLUTARIC ACID, DumPY: shorter than wild-type, ... | | Authors: | Brejc, K, Bian, Q, Uzawa, S, Wheeler, B.S, Anderson, E.C, King, D.S, Kranzusch, P.J, Preston, C.G, Meyer, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Dynamic Control of X Chromosome Conformation and Repression by a Histone H4K20 Demethylase.
Cell, 171, 2017
|
|
6M7K
 
 | | Structure of mouse RECON (AKR1C13) in complex with cyclic AMP-AMP-GMP (cAAG) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C13, cyclic AMP-AMP-GMP | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6MYD
 
 | | Structure of zebrafish TRAF6 in complex with STING CTT | | Descriptor: | STING CTT, Transmembrane protein 173, SULFATE ION, ... | | Authors: | de Oliveira Mann, C.C, Orzalli, M.H, King, D.S, Kagan, J.C, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Modular Architecture of the STING C-Terminal Tail Allows Interferon and NF-kappa B Signaling Adaptation.
Cell Rep, 27, 2019
|
|
6CTA
 
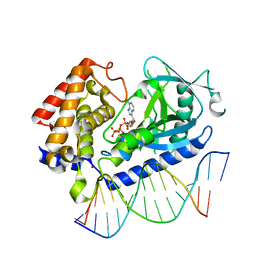 | | Structure of the human cGAS-DNA complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Zhou, W, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.779 Å) | | Cite: | Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance.
Cell, 174, 2018
|
|
6CT9
 
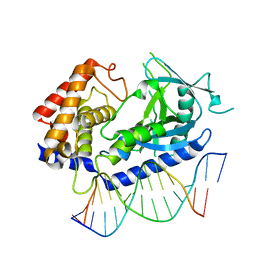 | | Structure of the human cGAS-DNA complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Zhou, W, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-03-22 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance.
Cell, 174, 2018
|
|
8U7I
 
 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
8SL0
 
 | |
4P6I
 
 | | Crystal structure of the Cas1-Cas2 complex from Escherichia coli | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Nunez, J.K, Kranzusch, P.J, Noeske, J, Doudna, J.A. | | Deposit date: | 2014-03-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity.
Nat.Struct.Mol.Biol., 21, 2014
|
|
