3KTZ
 
 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2POL
 
 | |
6TWB
 
 | |
6TWC
 
 | |
6W73
 
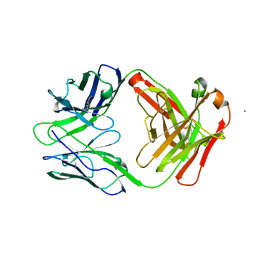 | |
2G35
 
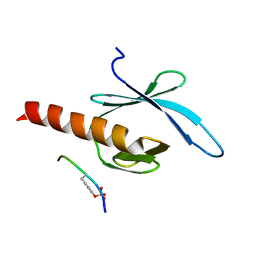 | | NMR structure of talin-PTB in complex with PIPKI | | Descriptor: | Talin-1, peptide | | Authors: | Kong, X, Wang, X, Misra, S, Qin, J. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Phosphorylation-regulated Focal Adhesion Targeting of Type Igamma Phosphatidylinositol Phosphate Kinase (PIPKIgamma) by Talin.
J.Mol.Biol., 359, 2006
|
|
3KU0
 
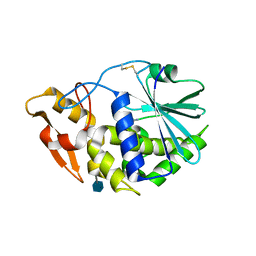 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
4XMK
 
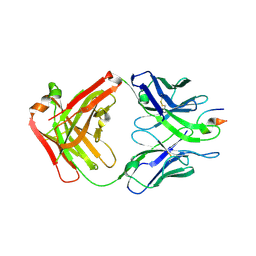 | |
3MLY
 
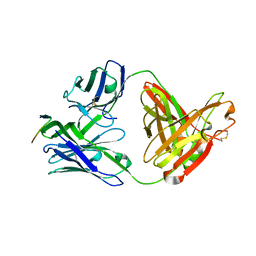 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a UR29 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLV
 
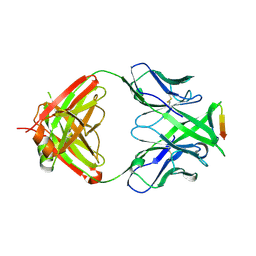 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with an NOF V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLW
 
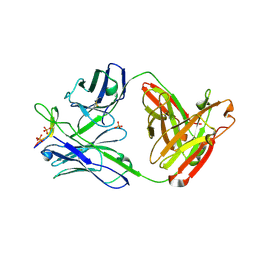 | | Crystal structure of anti-HIV-1 V3 Fab 1006-15D in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab light chain, ... | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLZ
 
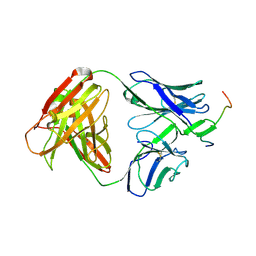 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a VI191 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLU
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a ZAM18 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLX
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLR
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a NY5 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLT
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a UG1033 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3UJI
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2558 in complex with MN peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein gp160, ... | | Authors: | Kong, X.P. | | Deposit date: | 2011-11-07 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Anti-V3 HIV-1 Monoclonal Antibodies Encoded by the VH5-51/VL Lambda Genes Define a Conserved Antigenic Structure.
Plos One, 6, 2011
|
|
3MLS
 
 | |
2F23
 
 | |
4XBX
 
 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBT
 
 | | Crystal Structure of the L74F/M78F/L103V/L114V/I116V/F139V/L147V mutant of LEH complexed with (S,S)-cyclohexanediol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, CITRATE ANION, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDV
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclohexanediol | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XBY
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XDW
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH | | Descriptor: | Limonene-1,2-epoxide hydrolase | | Authors: | Kong, X.D, Sun, Z, Lonsdale, R, Xu, J.H, Reetz, M.T, Zhou, J. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reshaping an Enzyme Binding Pocket for Enhanced and Inverted Stereoselectivity: Use of Smallest Amino Acid Alphabets in Directed Evolution
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3UJJ
 
 | |
