5AKP
 
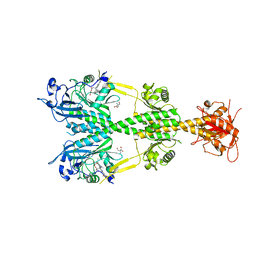 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
6PH2
 
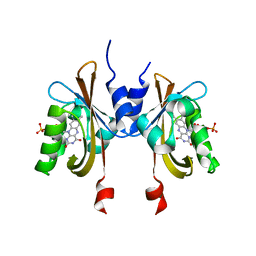 | | Complete LOV domain from the LOV-HK sensory protein from Brucella abortus (mutant C69S, construct 15-155) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
7L5A
 
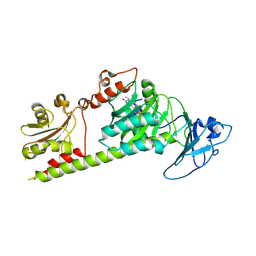 | | Crystal structure of the photosensory module from Xanthomonas campestris bacteriophytochrome XccBphP in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
7L59
 
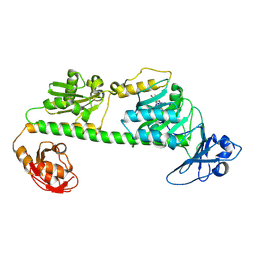 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP-G454E variant from Xanthomonas campestris in the Pfr state | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Antelo, G, Sanchez-Lamas, M, Klinke, S, Goldbaum, F.A, Rinaldi, J, Bonomi, H.R. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Sci Adv, 7, 2021
|
|
6PH3
 
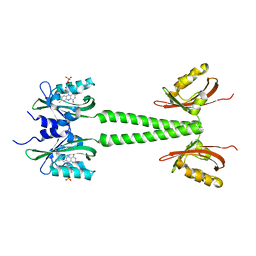 | | LOV-PAS construct from the LOV-HK sensory protein from Brucella abortus (dark-adapted, construct 15-273) | | Descriptor: | Blue-light-activated histidine kinase, FLAVIN MONONUCLEOTIDE | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
6ODD
 
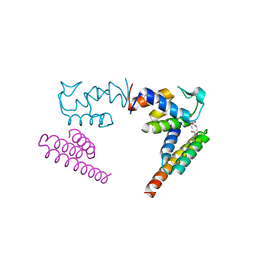 | | Crystal structure of the human complex ACP-ISD11 | | Descriptor: | Acyl carrier protein, mitochondrial, CALCIUM ION, ... | | Authors: | Herrera, M.G, Noguera, M.E, Klinke, S, Santos, J. | | Deposit date: | 2019-03-26 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Human ACP-ISD11 Heterodimer.
Biochemistry, 58, 2019
|
|
6OVP
 
 | |
5ANP
 
 | |
6UCT
 
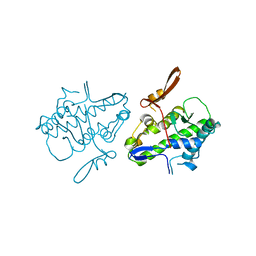 | | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Klinke, S, Monti, D, Sycz, G, Cerutti, M.L, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2019-09-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant)
To Be Published
|
|
6V95
 
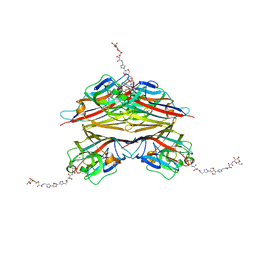 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-tartaramidoyl derivative (diNGT) | | Descriptor: | (2R,3R)-N-[(1-{(3S,3aR,6S,6aR)-6-[4-({[(2R,3R)-2,3-dihydroxy-4-oxo-4-{[(2R,3R,4R,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]amino}butanoyl]amino}methyl)-1H-1,2,3-triazol-1-yl]hexahydrofuro[3,2-b]furan-3-yl}-1H-1,2,3-triazol-4-yl)methyl]-2,3-dihydroxy-N'-[(2R,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]butanediamide (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-13 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VGF
 
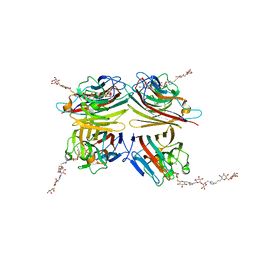 | | Peanut lectin complexed with divalent S-beta-D-thiogalactopyranosyl beta-D-glucopyranoside derivative (diSTGD) | | Descriptor: | (2S,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6R)-3,4,5-trihydroxy-6-{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({4-[({[(2S,3S,4S,5R,6S)-3,4,5-trihydroxy-6-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-2-yl]methyl}sulfanyl)methyl]-1H-1,2,3-triazol-1-yl}methyl)tetrahydro-2H-pyran-2-yl]oxy}tetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VAW
 
 | | Peanut lectin complexed with N-beta-D-galactopyranosyl-L-succinamoyl derivative (NGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VAV
 
 | | Peanut lectin complexed with divalent N-beta-D-galactopyranosyl-L-succinamoyl derivative (diNGS) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VC3
 
 | | Peanut lectin complexed with S-beta-D-thiogalactopyranosyl 6-deoxy-6-S-propynyl-beta-D-glucopyranoside (STG) | | Descriptor: | 6-S-(prop-2-yn-1-yl)-6-thio-beta-D-glucopyranosyl 1-thio-beta-D-galactopyranoside, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7KVC
 
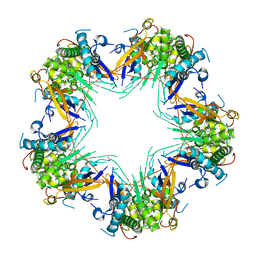 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7LRH
 
 | | C-terminal domain of RibD from Brucella abortus (5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5'-phosphate reductase) | | Descriptor: | Riboflavin biosynthesis protein RibD, SULFATE ION | | Authors: | Bonomi, H.R, Cerutti, M.L, Posadas, D.M, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2021-02-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-terminal domain of RibD from Brucella abortus (5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5'-phosphate reductase)
To be published
|
|
7MHW
 
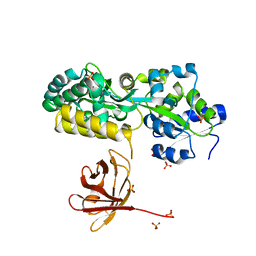 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
2Q76
 
 | | Mouse anti-hen egg white lysozyme antibody F10.6.6 Fab fragment | | Descriptor: | Fab F10.6.6 fragment Heavy Chain, Fab F10.6.6 fragment Light Chain | | Authors: | Cauerhff, A, Klinke, S, Acierno, J.P, Goldbaum, F.A, Braden, B.C. | | Deposit date: | 2007-06-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity maturation increases the stability and plasticity of the Fv domain of anti-protein antibodies.
J.Mol.Biol., 374, 2007
|
|
2QT7
 
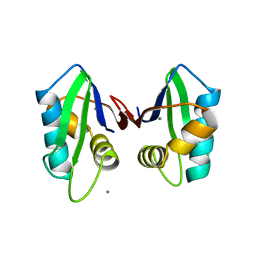 | | Crystallographic structure of the mature ectodomain of the human receptor-type protein-tyrosine phosphatase IA-2 at 1.30 Angstroms | | Descriptor: | CALCIUM ION, Receptor-type tyrosine-protein phosphatase-like N | | Authors: | Primo, M.E, Klinke, S, Sica, M.P, Goldbaum, F.A, Jakoncic, J, Poskus, E, Ermacora, M.R. | | Deposit date: | 2007-08-01 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the Mature Ectodomain of the Human Receptor-type Protein-tyrosine Phosphatase IA-2
J.Biol.Chem., 283, 2008
|
|
8DQT
 
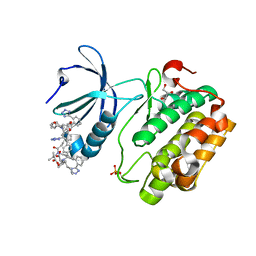 | | Human PDK1 kinase domain in complex with Valsartan | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL | | Authors: | Gross, L.Z.F, Klinke, S, Biondi, R.M. | | Deposit date: | 2022-07-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Modulation of the substrate specificity of the kinase PDK1 by distinct conformations of the full-length protein.
Sci.Signal., 16, 2023
|
|
8EK9
 
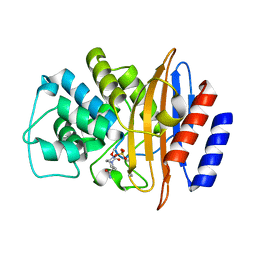 | | Crystal structure of the class A carbapenemase CRH-1 in complex with avibactam at 1.4 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
8EHU
 
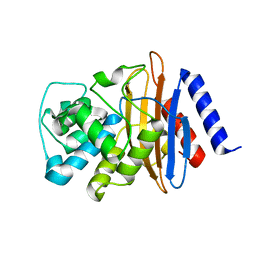 | | Crystal structure of the environmental CRH-1 class A carbapenemase at 1.1 Angstrom resolution | | Descriptor: | Beta-lactamase | | Authors: | Power, P, Brunetti, F, Ghiglione, B, Guardabassi, L, Gutkind, G, Klinke, S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and Structural Characterization of CRH-1, a Carbapenemase from Chromobacterium haemolyticum Related to KPC beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
8EO5
 
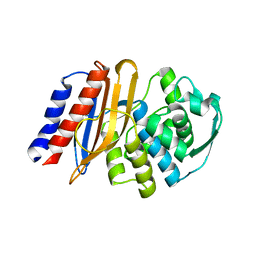 | | Crystal structure of the class A beta-lactamase precursor LRA-5 from an Alaskan soil metagenome at 1.8 Angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
