7Z68
 
 | | Pseudomonas aeruginosa Elastase in complex with a Thiol based inhibitor (R-and S-configured) | | Descriptor: | (2~{R})-~{N}-[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanylphenyl]-3-phenyl-2-sulfanyl-propanamide, CALCIUM ION, GLYCEROL, ... | | Authors: | Klein, A, Hirsch, A. | | Deposit date: | 2022-03-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual inhibitors of Pseudomonas aeruginosa virulence factors LecA and LasB
To Be Published
|
|
5EW5
 
 | |
6DGK
 
 | |
8RW3
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase, complexed with a non-covalent 1,2- Cyclophellitol aziridine | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, 1,2-ETHANEDIOL, OXALATE ION, ... | | Authors: | Moran, E, Davies, G, Ofamn, T, Heming, J, Nin-Hill, A, Kullmer, F, Steneker, R, Klein, A, Bennett, M, Ruijgrok, G, Kok, K, Aerts, J, Van der Marel, G, Rovira, C, Artola, M, Codee, J, Overkleeft, H. | | Deposit date: | 2024-02-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational and Electronic Variations in 1,2- and 1,5a-Cyclophellitols and their Impact on Retaining alpha-Glucosidase Inhibition.
Chemistry, 30, 2024
|
|
1N5Z
 
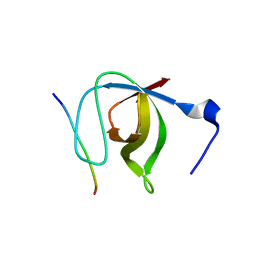 | | Complex structure of Pex13p SH3 domain with a peptide of Pex14p | | Descriptor: | 14-mer peptide from Peroxisomal membrane protein PEX14, Peroxisomal membrane protein PAS20 | | Authors: | Douangamath, A, Filipp, F.V, Klein, A.T.J, Barnett, P, Zou, P, Voorn-Brouwer, T, Vega, M.C, Mayans, O.M, Sattler, M, Distel, B, Wilmanns, M. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
MOL.CELL, 10, 2002
|
|
4JML
 
 | |
6U1N
 
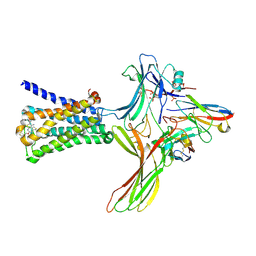 | | GPCR-Beta arrestin structure in lipid bilayer | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Staus, D.P, Hu, H, Robertson, M.J, Kleinhenz, A.L.W, Wingler, L.M, Capel, W.D, Latorraca, N.R, Lefkowitz, R.J, Skiniotis, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc.
Nature, 579, 2020
|
|
6OS0
 
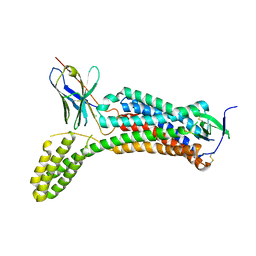 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to angiotensin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, CHLORIDE ION, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OS1
 
 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV023 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Nanobody Nb.AT110i1_le, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OS2
 
 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV026 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7Q
 
 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
4KNB
 
 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2N74
 
 | | Solution Structure of the RNA-Binding domain of non-structural protein 1 from the 1918 H1N1 influenza virus | | Descriptor: | Non-structural protein 1 | | Authors: | Jureka, A.S, Kleinpeter, A.B, Cornilescu, G, Cornilescu, C.C, Schwieters, C.D, Petit, C.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I.
Structure, 23, 2015
|
|
4ID7
 
 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
