5N34
 
 | | Thermolysin in complex with inhibitor JC276 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
5M69
 
 | | Thermolysin in complex with inhibitor and xenon | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-24 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
6SFA
 
 | |
5N31
 
 | | Thermolysin in complex with inhibitor JC277 | | Descriptor: | (2~{S})-6-azanyl-2-[[(2~{S})-4-methyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]pentanoyl]amino]hexanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.368 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
3BIV
 
 | | Human thrombin-in complex with UB-THR11 | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclohexylamino)ethanoyl)pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin, ... | | Authors: | Gerlach, C, Smolinski, M, Steuber, H, Sotriffer, C.A, Heine, A, Hangauer, D.G, Klebe, G. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic Inhibition Profile of a Cyclopentyl and a Cyclohexyl Derivative towards Thrombin: The Same but for Different Reasons
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
5LCR
 
 | |
6TXP
 
 | | Human Aldose Reductase Mutant L300A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | (2-carbamoyl-5-fluorophenoxy)acetic acid, 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Human Aldose Reductase Mutant L300A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
5M5F
 
 | | Thermolysin in complex with inhibitor and krypton | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-21 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
5N2Z
 
 | | Thermolysin in complex with inhibitor JC286 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Thermolysin, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N3V
 
 | | Thermolysin in complex with inhibitor JC292 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
6T5G
 
 | |
6TUF
 
 | | Human Aldose Reductase in complex with ALR43 | | Descriptor: | 2-[5-fluoranyl-2-[[3-[methyl(oxidanyl)-$l^{3}-sulfanyl]phenyl]methylcarbamoyl]phenoxy]ethanoic acid, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Human Aldose Reductase in complex with SAR25
To Be Published
|
|
5LPP
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group C2) with 6-amino-4-(2-((3aR,4R,6R,6aR)-6-methoxy-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)ethyl)-2-(methylamino)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 4-[2-[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-methoxy-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]ethyl]-6-azanyl-2-(methylamino)-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
5LWD
 
 | | Thermolysin in complex with inhibitor (JC96) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Elucidating the Origin of Long Residence Time Binding for Inhibitors of the Metalloprotease Thermolysin.
ACS Chem. Biol., 12, 2017
|
|
7AXT
 
 | |
7AXW
 
 | |
6T7Q
 
 | | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, CITRIC ACID, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2019-10-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Human Aldose Reductase Mutant L301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
6SF8
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase in complex with fragment j51 | | Descriptor: | 3-[(4E)-4-imino-5,6-dimethylfuro[2,3-d]pyrimidin-3(4H)-yl]-N,N-dimethylpropan-1-amine, Farnesyl diphosphate synthase, SULFATE ION | | Authors: | Magari, F, Heine, A, Klebe, G. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Trypanosoma cruzi farnesyl diphosphate synthase in complex with fragment j51
To Be Published
|
|
6SYW
 
 | | human Aldose Reductase in complex with SAR25 | | Descriptor: | 2-[2-(cyclopropylmethylcarbamoyl)-5-fluoranyl-phenoxy]ethanoic acid, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | human Aldose Reductase in complex with SAR25
To Be Published
|
|
5LIF
 
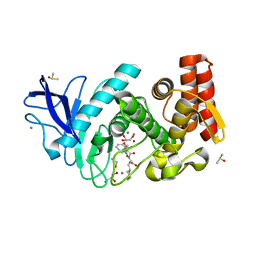 | | Thermolysin in complex with inhibitor | | Descriptor: | (2~{S})-3-cyclohexyl-2-[[(2~{S})-4-methyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]pentanoyl]amino]propanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Elucidating the Origin of Long Residence Time Binding for Inhibitors of the Metalloprotease Thermolysin.
ACS Chem. Biol., 12, 2017
|
|
6RSV
 
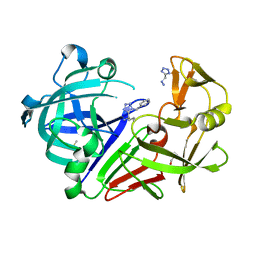 | | Endothiapepsin in complex with 017 | | Descriptor: | 1~{H}-1,2,3,4-tetrazol-5-ylmethyldiazane, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Magari, F, Heine, A, Klebe, G. | | Deposit date: | 2019-05-22 | | Release date: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Endothiapepsin in complex with 017
To Be Published
|
|
5LCQ
 
 | |
5LVD
 
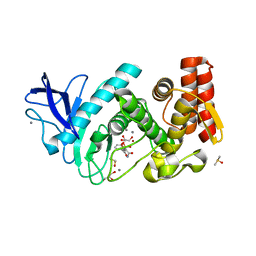 | | Thermolysin in complex with inhibitor (JC67) | | Descriptor: | (2~{S})-4-methyl-2-[[(2~{S})-3-oxidanyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]propanoyl]amino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-14 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
4FRC
 
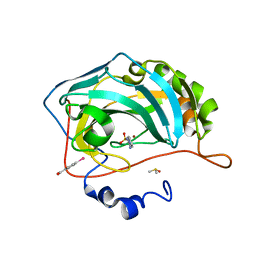 | |
5O6O
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal 2,6-pyridinketone inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, 2-fluoranyl-3-[6-(4-fluoranyl-3-oxidanyl-phenoxy)pyridin-2-yl]phenol, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Heine, A, Braun, F, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based design and profiling of novel 17 beta-HSD14 inhibitors.
Eur J Med Chem, 155, 2018
|
|
