3MDE
 
 | |
3MDD
 
 | |
6P8I
 
 | | N-terminal 5 domains of IGFIIR | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2019-06-07 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
6V02
 
 | | N-terminal 5 domains of CI-MPR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor | | Authors: | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
4XQM
 
 | |
4FVV
 
 | | Crystal structure of HCR/D-Sa-GBL1/C | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid, Neurotoxin, ... | | Authors: | Fu, Z, Karalewitz, A, Baldwin, M.R, Kim, J.-J.P, Barbieri, J.T. | | Deposit date: | 2012-06-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Botulinum neurotoxin serotype C associates with dual ganglioside receptors to facilitate cell entry.
J.Biol.Chem., 287, 2012
|
|
1XX4
 
 | | Crystal Structure of Rat Mitochondrial 3,2-Enoyl-CoA | | Descriptor: | 3,2-trans-enoyl-CoA isomerase, mitochondrial, BENZAMIDINE, ... | | Authors: | Hubbard, P.A, Yu, W, Schulz, H, Kim, J.-J. | | Deposit date: | 2004-11-03 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain swapping in the low-similarity isomerase/hydratase superfamily: the crystal structure of rat mitochondrial Delta3, Delta2-enoyl-CoA isomerase.
Protein Sci., 14, 2005
|
|
2RL9
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 6.5 bound to trimannoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Hindsgaul, O, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2007-10-18 | | Release date: | 2008-02-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of pH-dependent ligand binding and release by the cation-dependent mannose 6-phosphate receptor.
J.Biol.Chem., 283, 2008
|
|
2RL7
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 4.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CACODYLATE ION, ... | | Authors: | Olson, L.J, Hindsgaul, O, Kim, J.-J.P, Dahms, N.M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-02-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor.
J.Biol.Chem., 283, 2008
|
|
2RLB
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 6.5 bound to M6P in absence of Mn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor | | Authors: | Olson, L.J, Hindsgaul, O, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2007-10-18 | | Release date: | 2008-02-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor.
J.Biol.Chem., 283, 2008
|
|
2RL8
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 6.5 bound to M6P | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Hindsgaul, O, Kim, J.-J.P, Dahms, N.M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-02-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor.
J.Biol.Chem., 283, 2008
|
|
4IQP
 
 | | Crystal Structure of HCRA-W1266A | | Descriptor: | Botulinum neurotoxin type A, GLYCEROL | | Authors: | Fu, Z, Kroken, A.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2013-01-12 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancing the Protective Immune Response against Botulism.
Infect.Immun., 81, 2013
|
|
2CW6
 
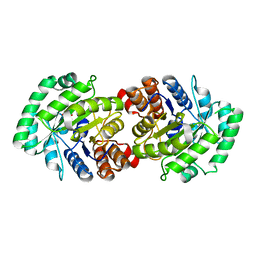 | | Crystal Structure of Human HMG-CoA Lyase: Insights into Catalysis and the Molecular Basis for Hydroxymethylglutaric Aciduria | | Descriptor: | 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, mitochondrial, ... | | Authors: | Fu, Z, Runquist, J.A, Hunt, J.F, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human 3-hydroxy-3-methylglutaryl-CoA Lyase: insights into catalysis and the molecular basis for hydroxymethylglutaric aciduria
J.Biol.Chem., 281, 2006
|
|
1JQI
 
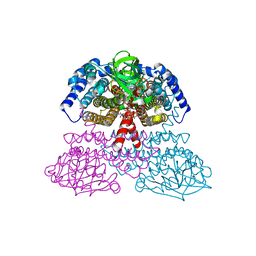 | | Crystal Structure of Rat Short Chain Acyl-CoA Dehydrogenase Complexed With Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, short chain acyl-CoA dehydrogenase | | Authors: | Battaile, K.P, Molin-Case, J, Paschke, R, Wang, M, Bennett, D, Vockley, J, Kim, J.-J.P. | | Deposit date: | 2001-08-07 | | Release date: | 2002-02-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases.
J.Biol.Chem., 277, 2002
|
|
3D4J
 
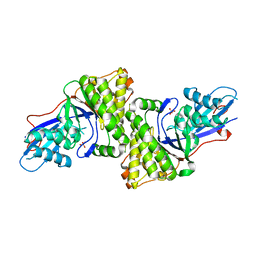 | | Crystal structure of Human mevalonate diphosphate decarboxylase | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Voynova, N.E, Fu, Z, Battaile, K, Herdendorf, T.J, Kim, J.-J.P, Miziorko, H.M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Arch.Biochem.Biophys., 480, 2008
|
|
3CY4
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 7.4 | | Descriptor: | Cation-dependent mannose-6-phosphate receptor, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Olson, L.J, Hindsgaul, O, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor.
J.Biol.Chem., 283, 2008
|
|
3ES9
 
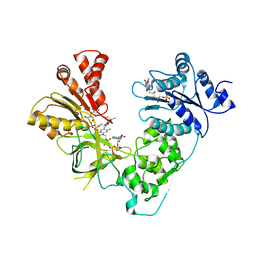 | | NADPH-Cytochrome P450 Reductase in an Open Conformation | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hamdane, D, Xia, C, Im, S.-C, Zhang, H, Kim, J.-J, Waskell, L. | | Deposit date: | 2008-10-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450
J.Biol.Chem., 284, 2009
|
|
3MP5
 
 | | Crystal Structure of Human Lyase R41M in complex with HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, Hydroxymethylglutaryl-CoA lyase, MAGNESIUM ION | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3MP4
 
 | | Crystal structure of Human lyase R41M mutant | | Descriptor: | Hydroxymethylglutaryl-CoA lyase | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3N7K
 
 | | Crystal structure of botulinum neurotoxin serotype C1 binding domain | | Descriptor: | Botulinum neurotoxin type C1 | | Authors: | Fu, Z, Kroken, A, Karalewitz, A, Baldwin, M.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a Unique Ganglioside Binding Loop within Botulinum Neurotoxins C and D-SA .
Biochemistry, 49, 2010
|
|
3N7J
 
 | | Crystal structure of botulinum neurotoxin serotype D binding domain | | Descriptor: | Botulinum neurotoxin type D | | Authors: | Fu, Z, Baldwin, M.R, Karalewitz, A, Kroken, A, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Unique Ganglioside Binding Loop within Botulinum Neurotoxins C and D-SA .
Biochemistry, 49, 2010
|
|
3N7M
 
 | | Crystal structure of W1252A mutant of HCR D/C VPI 5995 | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION | | Authors: | Fu, Z, Karalewitz, A, Kroken, A, Baldwin, M.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a Unique Ganglioside Binding Loop within Botulinum Neurotoxins C and D-SA .
Biochemistry, 49, 2010
|
|
3MP3
 
 | | Crystal Structure of Human Lyase in complex with inhibitor HG-CoA | | Descriptor: | (3R,5S,9R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, ... | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3N7L
 
 | | Crystal structure of botulinum neurotoxin serotype D/C VPI 5993 binding domain | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION | | Authors: | Fu, Z, Karalewitz, A, Kroken, A, Baldwin, M.R, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a Unique Ganglioside Binding Loop within Botulinum Neurotoxins C and D-SA .
Biochemistry, 49, 2010
|
|
2NYQ
 
 | |
