4HY8
 
 | | Structures of PR1 and PR2 intermediates from time-resolved laue crystallography | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Wulff, M, Moffat, K. | | 登録日 | 2012-11-13 | | 公開日 | 2013-03-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I38
 
 | | Structures of IT intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | 登録日 | 2012-11-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I3I
 
 | | Structures of IT intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected at 14ID APS | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | 登録日 | 2012-11-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
5XVC
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | 分子名称: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | 分子名称: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2019-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XVB
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | 分子名称: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | 著者 | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | 登録日 | 2017-06-27 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
1U5T
 
 | | Structure of the ESCRT-II endosomal trafficking complex | | 分子名称: | Defective in vacuolar protein sorting; Vps36p, Hypothetical 23.6 kDa protein in YUH1-URA8 intergenic region, appears to be functionally related to SNF7; Snf8p | | 著者 | Hierro, A, Sun, J, Rusnak, A.S, Kim, J, Prag, G, Emr, S.D, Hurley, J.H. | | 登録日 | 2004-07-28 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of ESCRT-II endosomal trafficking complex
Nature, 431, 2004
|
|
7VGX
 
 | | Neuropeptide Y Y1 Receptor (NPY1R) in Complex with G Protein and its endogeneous Peptide-Agonist Neuropeptide Y (NPY) | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Park, C, Kim, J, Jeong, H, Kang, H, Bang, I, Choi, H.-J. | | 登録日 | 2021-09-19 | | 公開日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of neuropeptide Y signaling through Y1 receptor
Nat Commun, 13, 2022
|
|
2KM9
 
 | |
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | 著者 | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | 登録日 | 2014-05-19 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
1Z3A
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Escherichia coli | | 分子名称: | ZINC ION, tRNA-specific adenosine deaminase | | 著者 | Malashkevich, V, Kim, J, Lisbin, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2005-03-10 | | 公開日 | 2006-02-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase.
Biochemistry, 45, 2006
|
|
1Y3G
 
 | | Crystal Structure of a Silanediol Protease Inhibitor Bound to Thermolysin | | 分子名称: | (2S)-2-{[(AMINOMETHYL)(DIHYDROXY)SILYL]METHYL}-4-METHYLPENTANAL, 3-PHENYLPROPANAL, CALCIUM ION, ... | | 著者 | Juers, D.H, Kim, J, Matthews, B.W, Sieburth, S.M. | | 登録日 | 2004-11-24 | | 公開日 | 2006-01-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Analysis of Silanediols as Transition-State-Analogue Inhibitors of the Benchmark Metalloprotease Thermolysin(,).
Biochemistry, 44, 2005
|
|
5U07
 
 | | CRISPR RNA-guided surveillance complex | | 分子名称: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | 著者 | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | 登録日 | 2016-11-23 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | 分子名称: | Protein tyrosine phosphatase type IVA 3 | | 著者 | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | 登録日 | 2013-07-29 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
3I4F
 
 | | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis | | 分子名称: | 3-oxoacyl-[acyl-carrier protein] reductase | | 著者 | Ramagopal, U.A, Kim, J, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-01 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure of putative 3-oxoacyl-reductase from bacillus thuringiensis
To be published
|
|
5CYU
 
 | | Structure of the soluble domain of EccB1 from the Mycobacterium smegmatis ESX-1 secretion system. | | 分子名称: | Conserved membrane protein | | 著者 | Arbing, M.A, Chan, S, Kahng, S, Kim, J, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2015-07-30 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system.
Bmc Struct.Biol., 16, 2016
|
|
3SJN
 
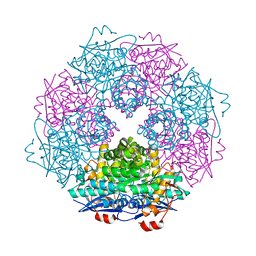 | | Crystal structure of enolase Spea_3858 (target EFI-500646) from Shewanella pealeana with magnesium bound | | 分子名称: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | 著者 | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-06-21 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Enolase Spea_3858 from Shewanella Pealeana
To be Published
|
|
5U0A
 
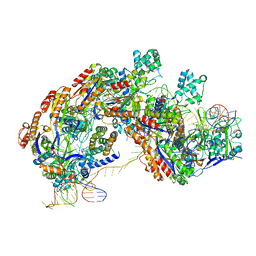 | | CRISPR RNA-guided surveillance complex | | 分子名称: | CRISPR-associated protein, Cas5e family, Cse1 family, ... | | 著者 | Xiao, Y, Luo, M, Hayes, R.P, Kim, J, Ng, S, Ding, F, Liao, M, Ke, A. | | 登録日 | 2016-11-23 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System.
Cell, 170, 2017
|
|
2OSC
 
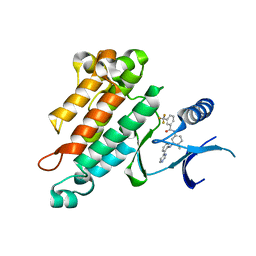 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | 分子名称: | Angiopoietin-1 receptor, N-{4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]PHENYL}-3-(TRIFLUOROMETHYL)BENZAMIDE | | 著者 | Bellon, S.F, Kim, J. | | 登録日 | 2007-02-05 | | 公開日 | 2007-03-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2OO8
 
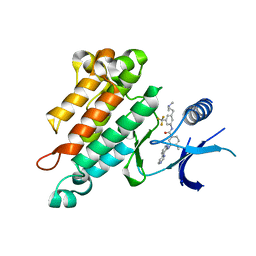 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | 分子名称: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | 著者 | Bellon, S.F, Kim, J. | | 登録日 | 2007-01-25 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3TTE
 
 | | Crystal structure of enolase brado_4202 (target EFI-501651) from Bradyrhizobium complexed with magnesium and mandelic acid | | 分子名称: | (S)-MANDELIC ACID, FORMIC ACID, GLYCEROL, ... | | 著者 | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-09-14 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Mandelate Racemase from Bradyrhizobium Sp. Ors278
To be Published
|
|
5IQ0
 
 | |
5IQ2
 
 | |
5IQ3
 
 | |
2KI2
 
 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | 分子名称: | Ss-DNA binding protein 12RNP2 | | 著者 | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | 登録日 | 2009-04-20 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
