1VFW
 
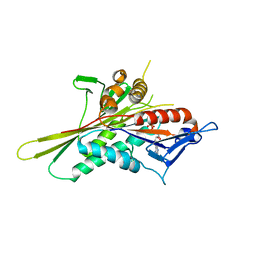 | | Crystal Structure of the Kif1A Motor Domain Complexed With Mg-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (Fusion protein consisting of Kinesin-like protein KIF1A, ... | | Authors: | Nitta, R, Kikkawa, M, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-04-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | KIF1A Alternately Uses Two Loops to Bind Microtubules
Science, 305, 2004
|
|
7D0I
 
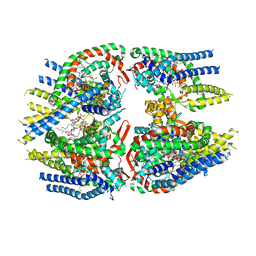 | | Cryo-EM structure of Schizosaccharomyces pombe Atg9 | | Descriptor: | Autophagy-related protein 9, Lauryl Maltose Neopentyl Glycol | | Authors: | Matoba, K, Tsutsumi, A, Kikkawa, M, Noda, N.N. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7EQ9
 
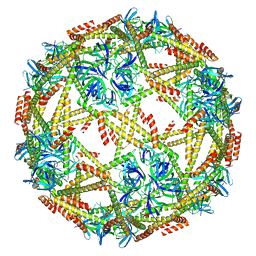 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | Descriptor: | TIP60 | | Authors: | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
7DCE
 
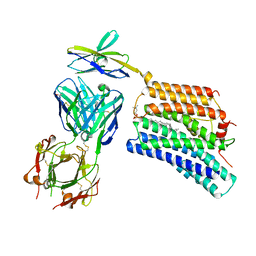 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8JUU
 
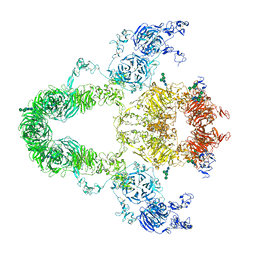 | | rat megalin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JX9
 
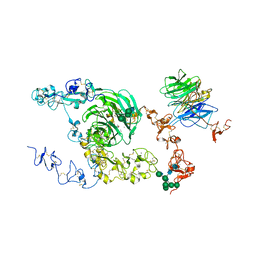 | | rat megalin bodyA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXH
 
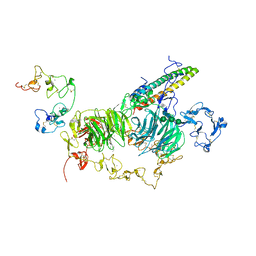 | | rat megalin RAP complex wingA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JX8
 
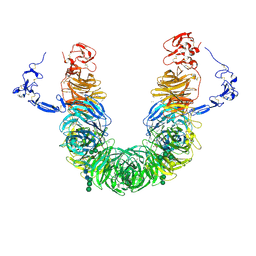 | | rat megalin head | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXF
 
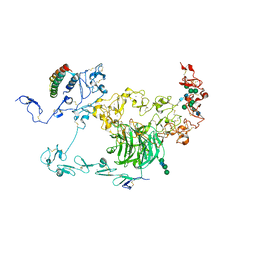 | | rat megalin RAP complex bodyA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin receptor-associated protein, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXJ
 
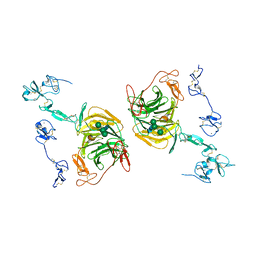 | | rat megalin RAP complex leg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LDL receptor related protein 2, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JUT
 
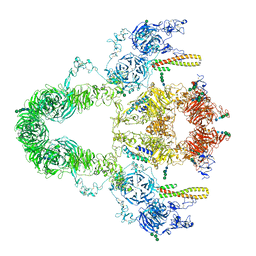 | | rat megalin RAP complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-27 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXE
 
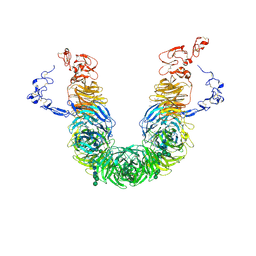 | | rat megalin RAP complex head | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXI
 
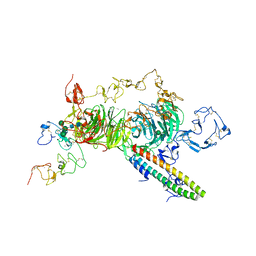 | | rat megalin RAP complex wingB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXG
 
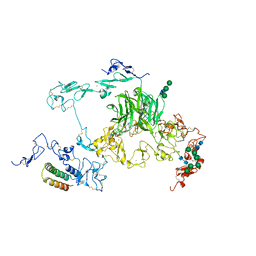 | | rat megalin RAP complex bodyB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXA
 
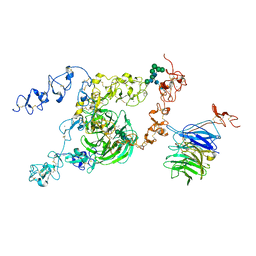 | | cryo-EM structure of rat megalin bodyB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXB
 
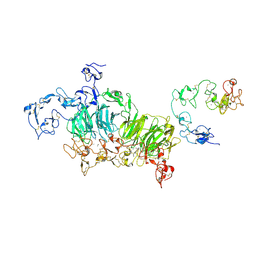 | | Cryo-EM structure of rat megalin wingAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXC
 
 | | rat megalin wingB | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JXD
 
 | | Cryo-EM structure of rat megalin leg | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto, S, Tsutsumi, A, Lee, Y, Hosojima, M, Kabasawa, H, Komochi, K, Yun-san, L, Nagatoshi, S, Tsumoto, K, Nishizawa, T, Kikkawa, M, Saito, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7VMR
 
 | | Structure of recombinant RyR2 mutant K4593A (EGTA dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMM
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMS
 
 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMN
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
