2VLN
 
 | | N75A mutant of E9 DNase domain in complex with Im9 | | 分子名称: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | 著者 | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | 登録日 | 2008-01-15 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
1F0K
 
 | | THE 1.9 ANGSTROM CRYSTAL STRUCTURE OF E. COLI MURG | | 分子名称: | SULFATE ION, UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE | | 著者 | Ha, S, Walker, D, Shi, Y, Walker, S. | | 登録日 | 2000-05-16 | | 公開日 | 2000-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis.
Protein Sci., 9, 2000
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | 分子名称: | De novo designed serine hydrolase, SODIUM ION | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6TC2
 
 | | Monoclinic human insulin in complex with p-coumaric acid | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Insulin, PHOSPHATE ION, ... | | 著者 | Triandafillidis, D.-P, Parthenios, N, Spiliopoulou, M, Valmas, A, Kosinas, C, Gozzo, F, Reinle-Schmitt, M, Beckers, D, Degen, T, Pop, M, Fitch, A, Wollenhaupt, J, Weiss, M.S, Karavassili, F, Margiolaki, I. | | 登録日 | 2019-11-04 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Insulin polymorphism induced by two polyphenols: new crystal forms and advances in macromolecular powder diffraction.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | 分子名称: | Lysozyme | | 著者 | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | 登録日 | 2011-04-26 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
4DRT
 
 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | 分子名称: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | 著者 | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-02-17 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
2A3J
 
 | | Structure of URNdesign, a complete computational redesign of human U1A protein | | 分子名称: | U1 small nuclear ribonucleoprotein A | | 著者 | Varani, G, Dobson, N, Dantas, G, Baker, D. | | 登録日 | 2005-06-24 | | 公開日 | 2006-06-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution Structural Validation of the Computational Redesign of Human U1A Protein
Structure, 14, 2006
|
|
6V9S
 
 | | Structure-based development of subtype-selective orexin 1 receptor antagonists | | 分子名称: | CHOLESTEROL, OLEIC ACID, Orexin receptor type 1,GlgA glycogen synthase chimera, ... | | 著者 | Hellmann, J, Drabek, M, Yin, J, Huebner, H, Kraus, F, Proell, T, Weikert, D, Kolb, P, Rosenbaum, D.M, Gmeiner, P. | | 登録日 | 2019-12-16 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure-based development of a subtype-selective orexin 1 receptor antagonist.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4L6R
 
 | | Structure of the class B human glucagon G protein coupled receptor | | 分子名称: | DI(HYDROXYETHYL)ETHER, Soluble cytochrome b562 and Glucagon receptor chimera | | 著者 | Siu, F.Y, He, M, de Graaf, C, Han, G.W, Yang, D, Zhang, Z, Zhou, C, Xu, Q, Wacker, D, Joseph, J.S, Liu, W, Lau, J, Cherezov, V, Katritch, V, Wang, M.W, Stevens, R.C, GPCR Network (GPCR) | | 登録日 | 2013-06-12 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of the human glucagon class B G-protein-coupled receptor.
Nature, 499, 2013
|
|
5TZO
 
 | | Computationally Designed Fentanyl Binder - Fen49*-Complex | | 分子名称: | CHLORIDE ION, Endo-1,4-beta-xylanase A, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | 著者 | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | 登録日 | 2016-11-22 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
5TVV
 
 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | 分子名称: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | 著者 | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | 登録日 | 2016-11-10 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-04-24 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.203 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | 分子名称: | OR187 | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-04-23 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9971 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
2LCB
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | 分子名称: | Lysozyme | | 著者 | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | 登録日 | 2011-04-26 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2L69
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR28 | | 分子名称: | Rossmann 2x3 fold protein | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Mao, A, Mao, B, Patel, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-11-17 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of de novo designed rossmann 2x3 fold protein, Northeast Structural Genomics Consortium Target OR28
To be Published
|
|
5TVY
 
 | | Computationally Designed Fentanyl Binder - Fen49 | | 分子名称: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Endo-1,4-beta-xylanase A | | 著者 | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | 登録日 | 2016-11-10 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
2MRA
 
 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459 | | 分子名称: | De novo designed protein OR459 | | 著者 | Pulavarti, S.V.S.R.K, Kipnis, Y, Sukumaran, D, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2014-07-02 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459
To be Published
|
|
2KS6
 
 | |
5GAJ
 
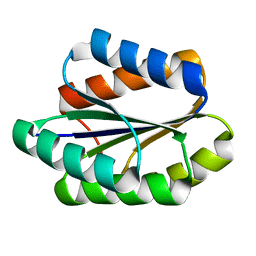 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | 分子名称: | DE NOVO DESIGNED PROTEIN OR258 | | 著者 | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
4EGG
 
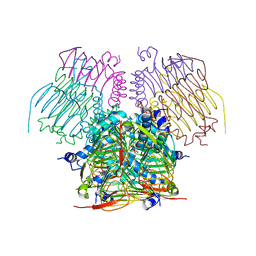 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | 分子名称: | GLYCEROL, Putative acetyltransferase SACOL2570 | | 著者 | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2012-03-30 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
2L9V
 
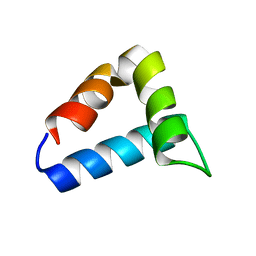 | | NMR structure of the FF domain L24A mutant's folding transition state | | 分子名称: | Pre-mRNA-processing factor 40 homolog A | | 著者 | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | 登録日 | 2011-02-24 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
8RGZ
 
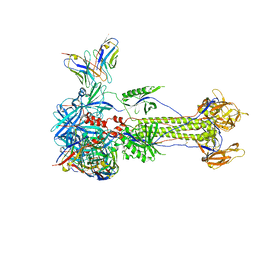 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | 分子名称: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | 著者 | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | 登録日 | 2023-12-14 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH0
 
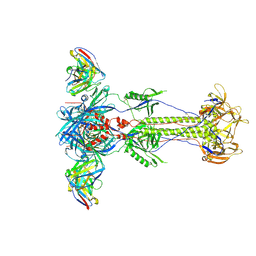 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | 分子名称: | Envelope glycoprotein B, HDIT102 Fab heavy chain | | 著者 | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | 登録日 | 2023-12-14 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
4JL8
 
 | |
4JLA
 
 | |
