4GLU
 
 | | Crystal structure of the mirror image form of VEGF-A | | Descriptor: | ACETATE ION, D- Vascular endothelial growth factor-A, GLYCEROL, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GLS
 
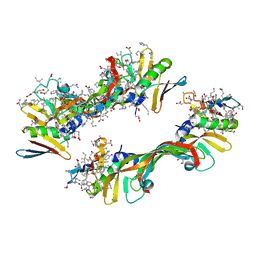 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4IUZ
 
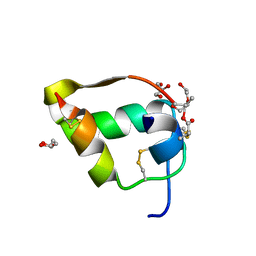 | | High resolution crystal structure of racemic ester insulin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin A chain, ... | | Authors: | Avital-Shmilovici, M, Mandal, K, Gates, Z.P, Phillips, N, Weiss, M.A, Kent, S.B.H. | | Deposit date: | 2013-01-22 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fully Convergent Chemical Synthesis of Ester Insulin: Determination of the High Resolution X-ray Structure by Racemic Protein Crystallography.
J.Am.Chem.Soc., 135, 2013
|
|
7LL8
 
 | | D-Protein RFX-V1 Bound to the VEGFR1 Domain 2 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V1 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7LL9
 
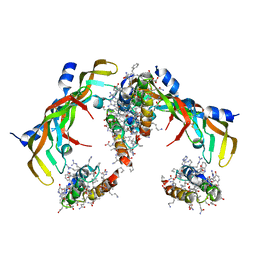 | | D-Protein RFX-V2 Bound to the VEGFR1 Domain 3 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V2 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
3HLO
 
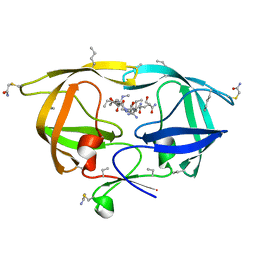 | |
3IAW
 
 | |
3KA2
 
 | |
3E8Y
 
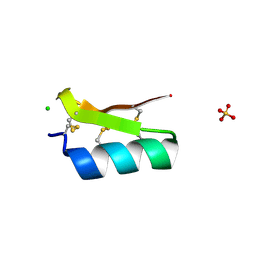 | | Xray structure of scorpion toxin BmBKTx1 | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 19.1, SULFATE ION | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-20 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structure of native scorpion toxin BmBKTx1 by racemic protein crystallography using direct methods.
J.Am.Chem.Soc., 131, 2009
|
|
3FSM
 
 | |
3E7R
 
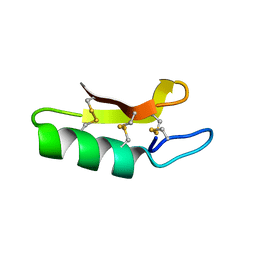 | | X-ray Crystal Structure of Racemic Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3E7U
 
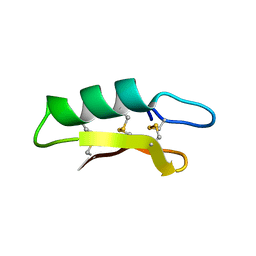 | | X-ray Crystal Structure of L-Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3HAU
 
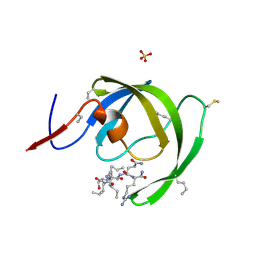 | |
3ODV
 
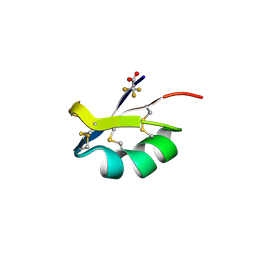 | | X-ray structure of kaliotoxin by racemic protein crystallography | | Descriptor: | CITRIC ACID, Potassium channel toxin alpha-KTx 3.1, trifluoroacetic acid | | Authors: | Pentelute, B.L, Mandal, K, Gates, Z.P, Sawaya, M.R, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-20 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Total chemical synthesis and X-ray structure of kaliotoxin by racemic protein crystallography.
Chem.Commun.(Camb.), 46, 2010
|
|
3NXE
 
 | |
3NGG
 
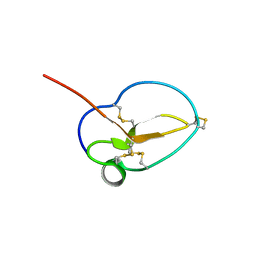 | | X-ray Structure of Omwaprin | | Descriptor: | Omwaprin-a | | Authors: | Banigan, J.R, Mandal, K, Sawaya, M.R, Thammavongsa, V, Hendrickx, A.P.A, Schneewind, O, Yeates, T.O, Kent, S.B.H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Protein Sci., 19, 2010
|
|
3NXN
 
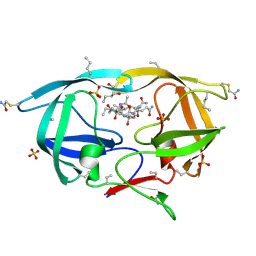 | | X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease covalent dimer | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-14 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NWX
 
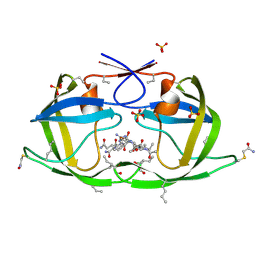 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-12 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NYG
 
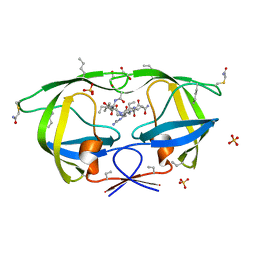 | |
3NWQ
 
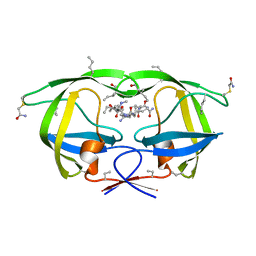 | |
3HZC
 
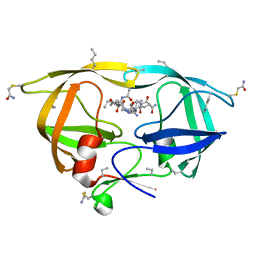 | |
3I2L
 
 | |
3IA9
 
 | |
5UDP
 
 | | High resolution x-ray crystal structure of synthetic insulin lispro | | Descriptor: | CHLORIDE ION, Insulin Lispro A chain, Insulin Lispro B chain, ... | | Authors: | Mandal, K, Dhayalan, B, Kent, S.B.H. | | Deposit date: | 2016-12-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | Scope and Limitations of Fmoc Chemistry SPPS-Based Approaches to the Total Synthesis of Insulin Lispro via Ester Insulin.
Chemistry, 23, 2017
|
|
3GI0
 
 | |
