1WR6
 
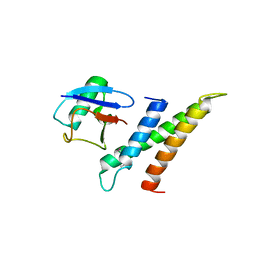 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1IO6
 
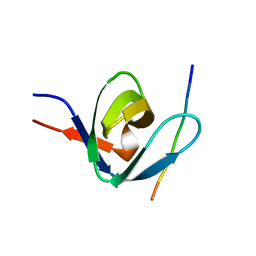 | |
7WAC
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) | | Descriptor: | Cyanophycin synthase | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
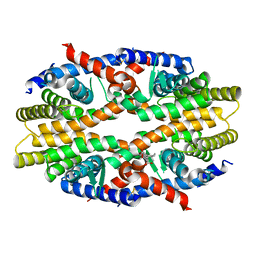
7WAD
 
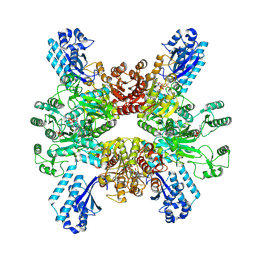 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS | | Descriptor: | Cyanophycin synthase, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
6JNR
 
 | | RXRa structure complexed with CU-6PMN and SRC1 peptide. | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RXRa structure complexed with CU-6PMN and SRC1 peptide.
To Be Published
|
|
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | Authors: | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | Deposit date: | 2019-11-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
4XKH
 
 | | CRYSTAL STRUCTURE OF THE AIRAPL TANDEM UIMS IN COMPLEX WITH A LYS48-LINKED TRI-UBIQUITIN | | Descriptor: | AN1-type zinc finger protein 2B, Polyubiquitin-C | | Authors: | Rahighi, S, Kawasaki, M, Stanhill, A, Wakatsuki, S. | | Deposit date: | 2015-01-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains.
Structure, 24, 2016
|
|
4DCN
 
 | | Crystal Structure Analysis of the Arfaptin2 BAR domain in Complex with ARL1 | | Descriptor: | ADP-ribosylation factor-like protein 1, Arfaptin-2, MAGNESIUM ION, ... | | Authors: | Nakamura, K, Xie, Y, Kawasaki, M, Kato, R, Wakatsuki, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for membrane binding specificity of the Bin/Amphiphysin/Rvs (BAR) domain of Arfaptin-2 determined by Arl1 GTPase
J.Biol.Chem., 287, 2012
|
|
4Z4M
 
 | |
4Z4K
 
 | |
8WCI
 
 | | Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae | | Descriptor: | CARDIOLIPIN, N,N-dimethyl-4-(5-methyl-1H-benzimidazol-2-yl)aniline, SODIUM ION, ... | | Authors: | Suzuki, K, Mikuriya, S, Adachi, N, Kawasaki, M, Senda, T, Moriya, T, Murata, T. | | Deposit date: | 2023-09-12 | | Release date: | 2024-10-09 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Na + -V-ATPase inhibitor curbs VRE growth and unveils Na + pathway structure.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8Y6I
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in nanodisc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent translocase ABCB1,mNeonGreen, CHOLESTEROL, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
1WRD
 
 | | Crystal structure of Tom1 GAT domain in complex with ubiquitin | | Descriptor: | Target of Myb protein 1, Ubiquitin | | Authors: | Akutsu, M, Kawasaki, M, Katoh, Y, Shiba, T, Yamaguchi, Y, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain.
Febs Lett., 579, 2005
|
|
5LRW
 
 | | Structure of Cezanne/OTUD7B OTU domain bound to ubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, Polyubiquitin-B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5LRV
 
 | | Structure of Cezanne/OTUD7B OTU domain bound to Lys11-linked diubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, PHOSPHATE ION, ... | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5LRX
 
 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5LRU
 
 | | Structure of Cezanne/OTUD7B OTU domain | | Descriptor: | OTU domain-containing protein 7B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
8Y6H
 
 | | P-glycoprotein in complex with UIC2 Fab and triple elacridar molecules in LMNG detergent | | Descriptor: | ATP-dependent translocase ABCB1,mNeonGreen, UIC2 Fab heavy chain, UIC2 Fab light chain, ... | | Authors: | Hamaguchi-Suzuki, N, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Anzai, N, Senda, T, Murata, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of P-glycoprotein bound to triple elacridar inhibitor molecules.
Biochem.Biophys.Res.Commun., 709, 2024
|
|
2IEZ
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8ZYO
 
 | | Cryo-EM Structure of astemizole-bound hERG Channel | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8ZYP
 
 | | Cryo-EM Structure of E-4031-bound hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2, ~{N}-[4-[1-[2-(6-methylpyridin-2-yl)ethyl]piperidin-4-yl]carbonylphenyl]methanesulfonamide | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
8ZYN
 
 | | Cryo-EM Structure of inhibitor-free hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 32, 2024
|
|
