4UM3
 
 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3920 | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
4UM1
 
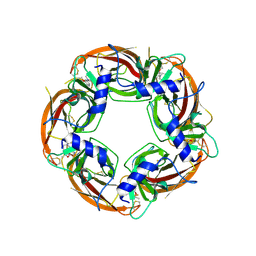 | | Engineered Ls-AChBP with alpha4-alpha4 binding pocket in complex with NS3573 | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Shahsavar, A, Kastrup, J.S, Balle, T, Gajhede, M. | | Deposit date: | 2014-05-14 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Achbp Engineered to Mimic the Alpha4-Alpha4 Binding Pocket in Alpha4Beta2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity
Mol.Pharmacol., 88, 2015
|
|
6HC9
 
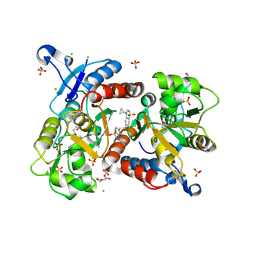 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 2.4 A RESOLUTION. | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
2CMO
 
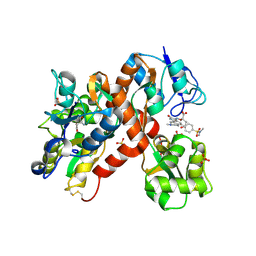 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | Descriptor: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
1RRU
 
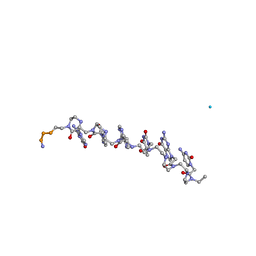 | | The influence of a chiral amino acid on the helical handedness of PNA in solution and in crystals | | Descriptor: | Peptide Nucleic Acid, (H-P(*CPN*GPN*TPN*APN*CPN*GPN)-LYS-NH2) | | Authors: | Rasmussen, H, Liljefors, T, Petersson, B, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
J.Biomol.Struct.Dyn., 21, 2004
|
|
1WVJ
 
 | | Exploring the GluR2 ligand-binding core in complex with the bicyclic AMPA analogue (S)-4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, GLYCEROL, SULFATE ION, ... | | Authors: | Nielsen, B.B, Pickering, D.S, Greenwood, J.R, Brehm, L, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the GluR2 ligand-binding core in complex with the bicyclical AMPA analogue (S)-4-AHCP
FEBS J., 272, 2005
|
|
2FPE
 
 | | Conserved dimerization of the ib1 src-homology 3 domain | | Descriptor: | C-jun-amino-terminal kinase interacting protein 1, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guenat, S, Dar, I, Bonny, C, Kastrup, J.S, Gajhede, M, Kristensen, O. | | Deposit date: | 2006-01-16 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unique set of SH3-SH3 interactions controls IB1 homodimerization
Embo J., 25, 2006
|
|
1XHY
 
 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | Authors: | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1XJ9
 
 | | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | | Descriptor: | peptide nucleic acid, (H-P(*GPN*TPN*APN*GPN*APN*TPN*CPN*APN*CPN*TPN)-LYS-NH2) | | Authors: | Petersson, B, Nielsen, B.B, Rasmussen, H, Larsen, I.K, Gajhede, M, Nielsen, P.E, Kastrup, J.S. | | Deposit date: | 2004-09-23 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network
J.Am.Chem.Soc., 127, 2005
|
|
3WIP
 
 | | Crystal structure of acetylcholine bound to Ls-AChBP | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholine-binding protein, ... | | Authors: | Olsen, J.A, Balle, T, Gajhede, M, Ahring, P.K, Kastrup, J.S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of the neurotransmitter acetylcholine by an acetylcholine binding protein reveals determinants of binding to nicotinic acetylcholine receptors
Plos One, 9, 2014
|
|
3ZDG
 
 | | Crystal Structure of Ls-AChBP complexed with carbamoylcholine analogue 3-(dimethylamino)butyl dimethylcarbamate (DMABC) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(dimethylamino)butyl dimethylcarbamate, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
4NWD
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | Descriptor: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4G8M
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist CBG-IV at 2.05A resolution | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
3GBA
 
 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | Descriptor: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
3GBB
 
 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with MSVIII-19 at 2.10A resolution | | Descriptor: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, Glutamate receptor, ionotropic kainate 1 | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
4ALX
 
 | | Crystal Structure of Ls-AChBP complexed with the potent nAChR antagonist DHbE | | Descriptor: | (4bS,6S)-6-methoxy-1,4,6,7,9,10,12,13-octahydro-3H,5H-pyrano[4',3':3,4]pyrido[2,1-i]indol-3-one, ACETYLCHOLINE BINDING PROTEIN, MAGNESIUM ION, ... | | Authors: | Shahsavar, A, Kastrup, J.S, Nielsen, E.O, Kristensen, J.L, Gajhede, M, Balle, T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lymnaea Stagnalis Achbp Complexed with the Potent Nachr Antagonist Dh-Betab-E Suggests a Unique Mode of Antagonism
Plos One, 7, 2012
|
|
2PBW
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
4H11
 
 | | Interaction partners of PSD-93 studied by X-ray crystallography and fluorescent polarization spectroscopy | | Descriptor: | ACETATE ION, Disks large homolog 2, SULFATE ION | | Authors: | Fiorentini, M, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Interaction partners of PSD-93 studied by X-ray crystallography and fluorescence polarization spectroscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZDH
 
 | | Crystal structure of Ls-AChBP complexed with carbamoylcholine analogue N,N-dimethyl-4-(1-methyl-1H-imidazol-2-yloxy)butan-2-amine | | Descriptor: | (2R)-N,N-dimethyl-4-(1-methylimidazol-2-yl)oxy-butan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
3ZRT
 
 | | Crystal structure of human PSD-95 PDZ1-2 | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Sorensen, P.L, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2011-06-19 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | A High-Affinity, Dimeric Inhibitor of Psd-95 Bivalently Interacts with Pdz1-2 and Protects Against Ischemic Brain Damage.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5MFW
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
5MFQ
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-344 at 1.90 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
4IGT
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution | | Descriptor: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2012-12-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
