3WNN
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
9CLY
 
 | | Crystal structure of the 3-ketoacyl-(acyl-carrier-protein) reductase, CylG, from Streptococcus agalactiae 2603V/R | | Descriptor: | CylG protein, IMIDAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-07-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the 3-ketoacyl-(acyl-carrier-protein) reductase, CylG, from Streptococcus agalactiae 2603V/R
To be published
|
|
7BU2
 
 | | Structure of alcohol dehydrogenase YjgB from Escherichia coli | | Descriptor: | Alcohol dehydrogenase, GLYCEROL, NITRATE ION, ... | | Authors: | Nguyen, G.T, Kim, Y.-G, Ahn, J.-W, Chang, J.H. | | Deposit date: | 2020-04-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli .
Molecules, 25, 2020
|
|
6CSM
 
 | | Crystal structure of the natural light-gated anion channel GtACR1 | | Descriptor: | GtACR1, OLEIC ACID, RETINAL | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
4UY5
 
 | |
4UY7
 
 | |
4UY6
 
 | |
3ZGA
 
 | | Crystal Structure of Penicillin-Binding Protein 4 from Listeria monocytogenes in the Carbenicillin bound form | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, PENICILLIN-BINDING PROTEIN 4 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structures of Bifunctional Penicillin-Binding Protein 4 from Listeria Monocytogenes.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZPJ
 
 | | Crystal structure of Ton1535 from Thermococcus onnurineus NA1 | | Descriptor: | TON_1535 | | Authors: | Jeong, J.H, kim, Y.G. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structure of the Hypothetical Protein Ton1535 from Thermococcus Onnurineus Na1 Reveals Unique Structural Properties by a Left-Handed Helical Turn in Normal Alpha-Solenoid Protein.
Proteins, 82, 2014
|
|
3ZG7
 
 | |
1T51
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1T55
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-02 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1T52
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-01 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
5UZC
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P221 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P221
To Be Published
|
|
4R3Z
 
 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
6J10
 
 | | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly | | Descriptor: | 6-cyclohexyl-4-methyl-1-oxidanyl-pyridin-2-one, Capsid protein | | Authors: | Park, S, Jin, M.S, Cho, Y, Kang, J, Kim, S, Park, M, Park, H, Kim, J, Park, S, Hwang, J, Kim, Y, Kim, Y.J. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ciclopirox inhibits Hepatitis B Virus secretion by blocking capsid assembly.
Nat Commun, 10, 2019
|
|
5V0I
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, TRYPTOPHAN, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Grimshaw, S.G, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan
To Be Published
|
|
5DGJ
 
 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
5DG6
 
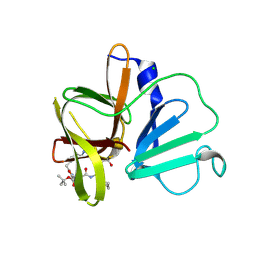 | | 2.35A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (21-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, CHLORIDE ION, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-23-oxa-6,9,14,21,22-pentaazabicyclo[18.2.1]tricosa-1(22),20-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
3ZG9
 
 | |
6VYO
 
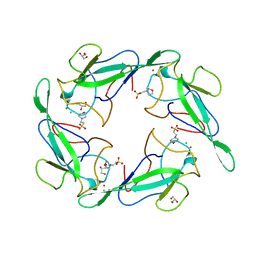 | | Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
5KZV
 
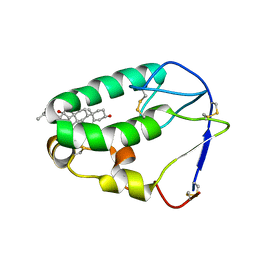 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in complex with 20(S)-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Smoothened | | Authors: | Huang, P, Kim, Y, Salic, A. | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.616 Å) | | Cite: | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
7O2L
 
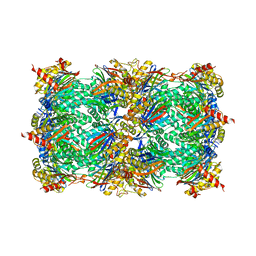 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
