5CI7
 
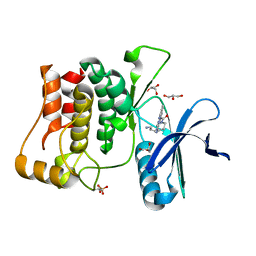 | | Structure of ULK1 bound to a selective inhibitor | | Descriptor: | GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]-5-iodopyrimidin-2-yl}amino)phenyl]pyrrolidine-1-carboxamide, Serine/threonine-protein kinase ULK1 | | Authors: | Lazarus, M.B, Shokat, K.M. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and structure of a new inhibitor scaffold of the autophagy initiating kinase ULK1.
Bioorg.Med.Chem., 23, 2015
|
|
5CE8
 
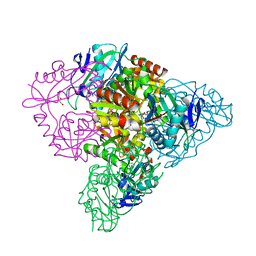 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis | | Descriptor: | Branched-chain amino acid aminotransferase, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for L-amino acids and R-amines.
Extremophiles, 20, 2016
|
|
5CM0
 
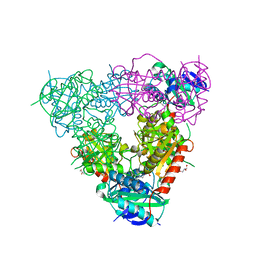 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans | | Descriptor: | Branched-chain transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5CRH
 
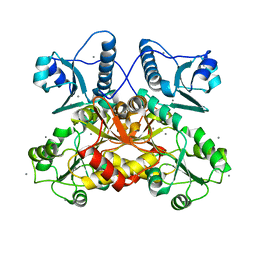 | |
5CRE
 
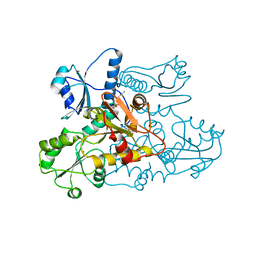 | | Human skeletal calsequestrin, D210G mutant low-calcium complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.315 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
5D1U
 
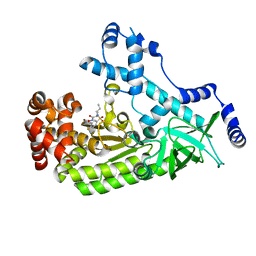 | | Anthrax toxin lethal factor with hydroxamic acid inhibitor | | Descriptor: | Lethal factor, N~2~-(6-aminohexyl)-N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ZINC ION | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2015-08-04 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | Probing the S2' Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates.
J.Med.Chem., 58, 2015
|
|
5D4H
 
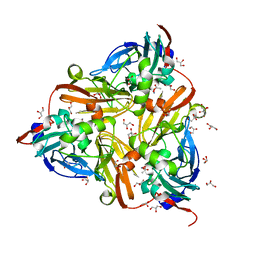 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CRD
 
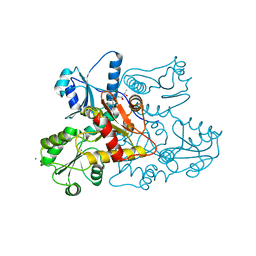 | | Wild-type human skeletal calsequestrin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calsequestrin-1 | | Authors: | Lewis, K.M, Ronish, L.A, Kang, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization of Two Human Skeletal Calsequestrin Mutants Implicated in Malignant Hyperthermia and Vacuolar Aggregate Myopathy.
J.Biol.Chem., 290, 2015
|
|
5DG5
 
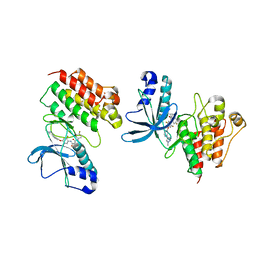 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
5DZX
 
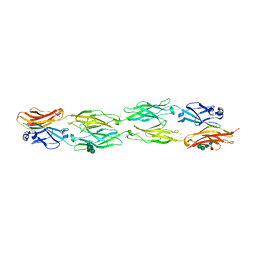 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5E9N
 
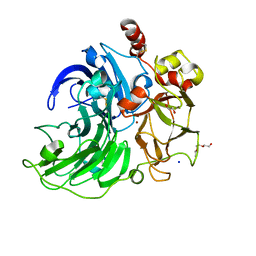 | | Steccherinum murashkinskyi laccase at 0.95 resolution | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Glazunova, O.A, Fedorova, T.V, Koroleva, O.V. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi.
Int. J. Biol. Macromol., 118, 2018
|
|
3MDC
 
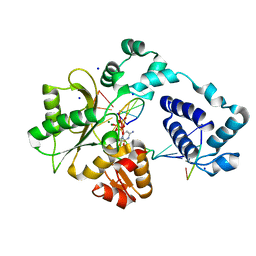 | | DNA polymerase lambda in complex with dFdCTP | | Descriptor: | 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*GP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Garcia-Diaz, M, Murray, M, Kunkel, T, Chou, K.M. | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Interaction between DNA Polymerase lambda and anticancer nucleoside analogs.
J.Biol.Chem., 285, 2010
|
|
5DZW
 
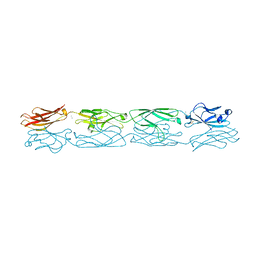 | | Protocadherin alpha 4 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin alpha-4, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
5DZV
 
 | | Protocadherin alpha 7 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein Pcdha7, ... | | Authors: | Goodman, K.M, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
3N59
 
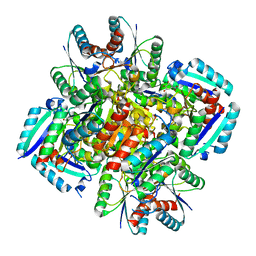 | | Type II dehydroquinase from Mycobacterium Tuberculosis complexed with 3-dehydroshikimate | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N7U
 
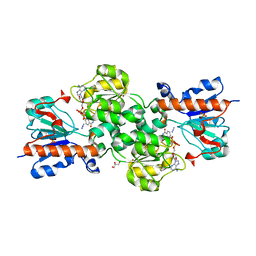 | | NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana in complex with NAD and azide | | Descriptor: | AZIDE ION, Formate dehydrogenase, GLYCEROL, ... | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
To be Published
|
|
3N86
 
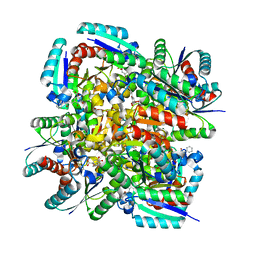 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 4 | | Descriptor: | (1R,5R)-1,5-dihydroxy-4-oxo-3-[3-oxo-3-(phenylamino)propyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
5ENP
 
 | | MBX2931 bound structure of bacterial efflux pump. | | Descriptor: | 6-[2-(3,4-dimethoxyphenyl)ethylsulfanyl]-8-[4-(2-methoxyethyl)piperazin-1-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridine-5-carbonitrile, DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3N76
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with compound 5 | | Descriptor: | (1S,3R,4R,5S)-1,3,4-TRIHYDROXY-5-(3-PHENOXYPROPYL)CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
5EQ4
 
 | | Crystal structure of the SrpA adhesin R347E mutant from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
3NMH
 
 | | Crystal structure of the abscisic receptor PYL2 in complex with pyrabactin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5D4J
 
 | | Chloride-bound form of a copper nitrite reductase from Alcaligenes faecals | | Descriptor: | ACETIC ACID, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ENQ
 
 | | MBX3132 bound structure of bacterial efflux pump. | | Descriptor: | DARPin, Multidrug efflux pump subunit AcrB,Multidrug efflux pump subunit AcrB, ~{N}-[4-[2-[[5-cyano-8-[(2~{S},6~{R})-2,6-dimethylmorpholin-4-yl]-3,3-dimethyl-1,4-dihydropyrano[3,4-c]pyridin-6-yl]sulfanyl]ethyl]phenyl]ethanamide | | Authors: | Sjuts, H, Ornik, A.R, Pos, K.M. | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3N87
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 3 | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[3-(phenylcarbonyl)phenyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
