4OSG
 
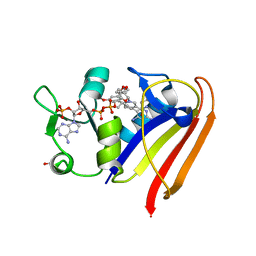 | | Klebsiella pneumoniae complexed with NADPH and 6-ethyl-5-[(3R)-3-[3-methoxyl-5-(pyridine-4-yl)phenyl]but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP1006) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Klebsiella pneumoniae Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal Features for Potency and Selectivity.
Antimicrob.Agents Chemother., 58, 2014
|
|
8ENT
 
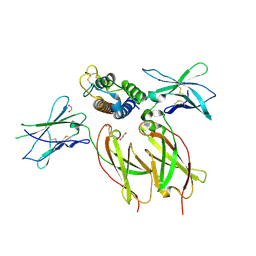 | | Interleukin-21 signaling complex with IL-21R and IL-2Rg | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abhiraman, G.C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A structural blueprint for interleukin-21 signal modulation.
Cell Rep, 42, 2023
|
|
8EDV
 
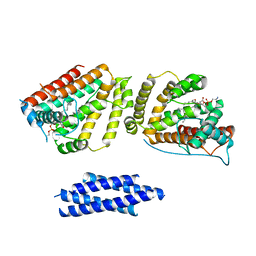 | |
4PAY
 
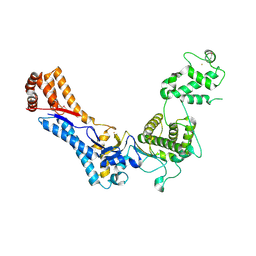 | | Crystal structure of an N-terminal fragment of the Legionella pneumophila effector protein SidC. | | Descriptor: | BARIUM ION, SidC, interaptin | | Authors: | Horenkamp, F.A, Mukherjee, S, Alix, E, Schauder, C.M, Hubber, A.M, Roy, C.R, Reinisch, K.M. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Legionella pneumophila Subversion of Host Vesicular Transport by SidC Effector Proteins.
Traffic, 15, 2014
|
|
4PKR
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 10 | | Descriptor: | CHLORIDE ION, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P53
 
 | | ValA (2-epi-5-epi-valiolone synthase) from Streptomyces hygroscopicus subsp. jinggangensis 5008 with NAD+ and Zn2+ bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Cyclase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kean, K.M, Codding, S.J, Karplus, P.A. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a sedoheptulose 7-phosphate cyclase: ValA from Streptomyces hygroscopicus.
Biochemistry, 53, 2014
|
|
4PKS
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NYA
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with 5-(azidomethyl)-2-methylpyrimidin-4-amine | | Descriptor: | 5-(azidomethyl)-2-methylpyrimidin-4-amine, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
4O44
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ529), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[3-(morpholin-4-yl)propoxy]-6-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
4O4G
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with 4-((4-(mesitylamino)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ527), a non-nucleoside inhibitor | | Descriptor: | 4-({4-[(2,4,6-trimethylphenyl)amino]-1,3,5-triazin-2-yl}amino)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Mislak, A.C, Frey, K.M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of NNRTIs targeting HIV-1 reverse transcriptase.
Biochim.Biophys.Acta, 1840, 2014
|
|
8EE1
 
 | | KS-AT didomain from module 2 of the 6-deoxyerythronolide B synthase in complex with antibody fragment AA5 | | Descriptor: | 6-deoxyerythronolide B synthase, AA5 antibody heavy chain, AA5 antibody light chain | | Authors: | Cogan, D.P, Brodsky, K.L, Guzman, K.M, Mathews, I.I, Khosla, C. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Biochemistry, 62, 2023
|
|
8EE0
 
 | | KS-AT didomain from module 2 of the 6-deoxyerythronolide B synthase in complex with antibody fragment 1B2 | | Descriptor: | 1B2 antibody heavy chain, 1B2 antibody light chain, 6-deoxyerythronolide B synthase | | Authors: | Cogan, D.P, Brodsky, K.L, Guzman, K.M, Mathews, I.I, Khosla, C. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and Characterization of Antibody Probes of Module 2 of the 6-Deoxyerythronolide B Synthase.
Biochemistry, 62, 2023
|
|
8EFN
 
 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
4NEU
 
 | | X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor | | Descriptor: | 1-[4-(1-aminoisoquinolin-5-yl)phenyl]-3-(5-tert-butyl-1,2-oxazol-3-yl)urea, MAGNESIUM ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Nolte, R.T, Ward, P, kahler, K.M, Campobasso, N. | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis.
ACS Med Chem Lett, 4, 2013
|
|
4NUF
 
 | | Crystal Structure of SHP/EID1 | | Descriptor: | EID1 peptide, Maltose ABC transporter periplasmic protein, Nuclear receptor subfamily 0 group B member 2 chimeric construct, ... | | Authors: | Zhi, X, Zhou, X.E, He, Y, Zechner, C, Suino-Powell, K.M, Kliewer, S.A, Melcher, K, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into gene repression by the orphan nuclear receptor SHP.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PKV
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 16 | | Descriptor: | Lethal factor, N~2~-[4-(aminomethyl)benzyl]-N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ZINC ION | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PGH
 
 | | Caffeic acid O-methyltransferase from Sorghum bicolor | | Descriptor: | Caffeic acid O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Green, A.R, Lewis, K.M, Kang, C. | | Deposit date: | 2014-05-02 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations.
Plant Physiol., 165, 2014
|
|
4PKT
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 13 | | Descriptor: | Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-N~2~-(4-nitrobenzyl)-D-alaninamide, ZINC ION | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8FFE
 
 | | Crystal structure of LRP6 E1E2 domains bound to YW210.09 Fab and engineered XWnt8 peptide | | Descriptor: | GLYCEROL, Low-density lipoprotein receptor-related protein 6, SODIUM ION, ... | | Authors: | Jude, K.M, Tsutsumi, N, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of the Wnt-Frizzled-LRP6 initiation complex reveals the basis for coreceptor discrimination.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4PGG
 
 | | Caffeic acid O-methyltransferase from Sorghum bicolor | | Descriptor: | Caffeic acid O-methyltransferase | | Authors: | Green, A.R, Lewis, K.M, Kang, C. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations.
Plant Physiol., 165, 2014
|
|
8FYA
 
 | | Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-containing prespacer complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (28-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FYB
 
 | | Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (17-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FY9
 
 | | Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-deficient prespacer complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (28-MER) | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FYD
 
 | | Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex bent CRISPR repeat conformation | | Descriptor: | Cas1, Cas2-DEDDh, DNA (13-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
