5L2F
 
 | | High Resolution Structure of Acinetobacter baumannii beta-lactamase OXA-51 I129L/K83D bound to doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ACETATE ION, Beta-lactamase, ... | | Authors: | June, C.M, Powers, R.A, Leonard, D.A, Muckenthaler, T. | | Deposit date: | 2016-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity.
Protein Sci., 25, 2016
|
|
5KZH
 
 | |
4MLL
 
 | | The 1.4 A structure of the class D beta-lactamase OXA-1 K70D complexed with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase OXA-1, ... | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2013-09-06 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural Origins of Oxacillinase Specificity in Class D beta-Lactamases.
Antimicrob.Agents Chemother., 58, 2014
|
|
4F94
 
 | | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Beta-lactamase, SULFATE ION | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2012-05-18 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin
To be Published
|
|
1HIP
 
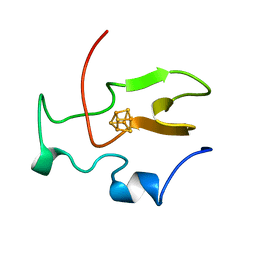 | | TWO-ANGSTROM CRYSTAL STRUCTURE OF OXIDIZED CHROMATIUM HIGH POTENTIAL IRON PROTEIN | | Descriptor: | HIGH POTENTIAL IRON PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Carterjunior, C.W, Kraut, J, Freer, S.T, Xuong, N.-H, Alden, R.A, Bartsch, R.G. | | Deposit date: | 1975-04-01 | | Release date: | 1976-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein.
J.Biol.Chem., 249, 1974
|
|
3QD2
 
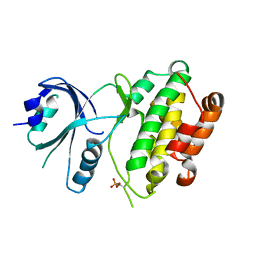 | | Crystal structure of mouse PERK kinase domain | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wenjun, C, Jingzhi, L, David, R, Bingdong, S. | | Deposit date: | 2011-01-17 | | Release date: | 2011-04-27 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of the PERK kinase domain suggests the mechanism for its activation.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5I2L
 
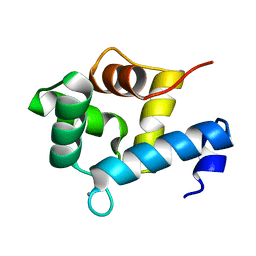 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
5I2Q
 
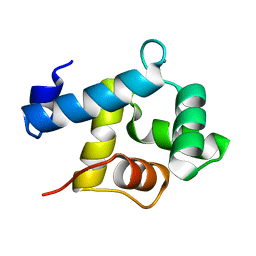 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
5I2O
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
7CLT
 
 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
5H0P
 
 | | Crystal structure of EF-hand protein mutant | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, An, J.Y, Kang, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Mun, S.A, Jun, C.D, Song, W.K, Eom, S.H. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Structural mechanism underlying regulation of human EFhd2/Swiprosin-1 actin-bundling activity by Ser183 phosphorylation.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQA
 
 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MJN
 
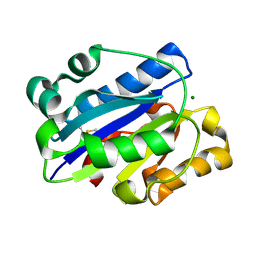 | | Crystal Structure of the intermediate affinity aL I domain mutant | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1P53
 
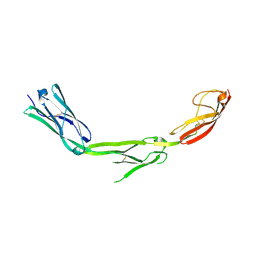 | | The Crystal Structure of ICAM-1 D3-D5 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1 | | Authors: | Yang, Y, Jun, C.D, Liu, J.H, Zhang, R, Jochimiak, A, Springer, T.A, Wang, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis for dimerization of ICAM-1 on the cell surface.
Mol.Cell, 14, 2004
|
|
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | Descriptor: | Beta-amyloid, Split Green flourescent protein | | Authors: | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
4X53
 
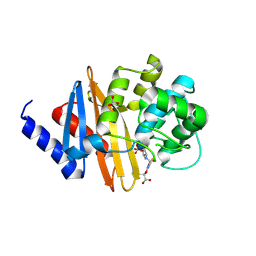 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BICARBONATE ION, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4X55
 
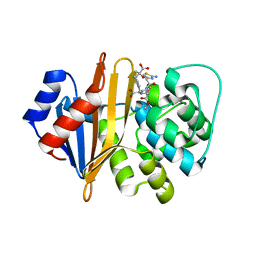 | | Structure of the class D Beta-Lactamase OXA-225 K82D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-225 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4X56
 
 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
8FQM
 
 | | ADC-7 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, GLYCINE | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fernando, M.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
8FQT
 
 | | Apo ADC-219 beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Maurer, O.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
8FQO
 
 | | ADC-33 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fish, E.R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
8FQN
 
 | | apo ADC-33 beta-lactamase | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fish, E.R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.256 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
8FQP
 
 | | apo ADC-162 beta-lactamase | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fernando, M.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
