7OCB
 
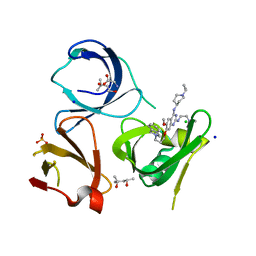 | | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[3-(1,3-dihydroisoindol-2-yl)propoxy]-2N-[2-(dimethylamino)ethyl]-6-methoxy-4N-(1-propan-2-ylpiperidin-4-yl)quinazoline-2,4-diamine, CHLORIDE ION, ... | | Authors: | Johansson, C, Krojer, T, Park, K, Xiong, Y, Jin, J, Oppermann, U. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B
To Be Published
|
|
7BBU
 
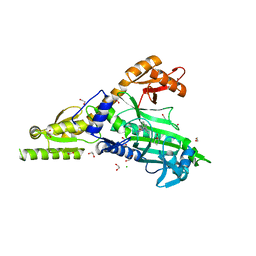 | | Crystal Structure of human Prolyl-tRNA synthetase in complex with NCP26 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional glutamate/proline--tRNA ligase, CHLORIDE ION, ... | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Krojer, T, Oppermann, U.C.T. | | Deposit date: | 2020-12-18 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of human Prolyl-tRNA synthetase in complex with NCP26 and L-Proline
To Be Published
|
|
1BOC
 
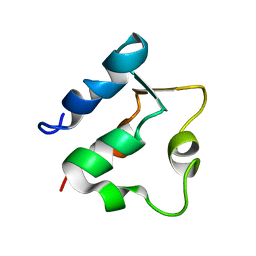 | | THE SOLUTION STRUCTURES OF MUTANT CALBINDIN D9K'S, AS DETERMINED BY NMR, SHOW THAT THE CALCIUM BINDING SITE CAN ADOPT DIFFERENT FOLDS | | Descriptor: | CALBINDIN D9K | | Authors: | Johansson, C, Ullner, M, Drakenberg, T. | | Deposit date: | 1993-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Biochemistry, 32, 1993
|
|
1BOD
 
 | | THE SOLUTION STRUCTURES OF MUTANT CALBINDIN D9K'S, AS DETERMINED BY NMR, SHOW THAT THE CALCIUM BINDING SITE CAN ADOPT DIFFERENT FOLDS | | Descriptor: | CALBINDIN D9K | | Authors: | Johansson, C, Ullner, M, Drakenberg, T. | | Deposit date: | 1993-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The solution structures of mutant calbindin D9k's, as determined by NMR, show that the calcium-binding site can adopt different folds.
Biochemistry, 32, 1993
|
|
6T7K
 
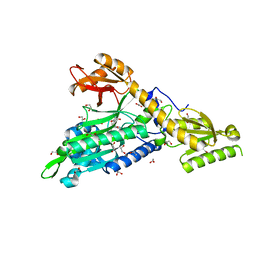 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Wang, J, Tye, M, Payne, N.C, Mazitschek, R, Thompson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline
To Be Published
|
|
1RKJ
 
 | | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target | | Descriptor: | 5'-R(*GP*GP*AP*UP*GP*CP*CP*UP*CP*CP*CP*GP*AP*GP*UP*GP*CP*AP*UP*CP*C)-3', Nucleolin | | Authors: | Johansson, C, Finger, L.D, Trantirek, L, Mueller, T.D, Kim, S, Laird-Offringa, I.A, Feigon, J. | | Deposit date: | 2003-11-21 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target.
J.Mol.Biol., 337, 2004
|
|
7QB7
 
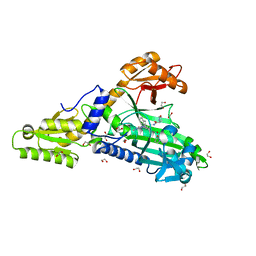 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT345 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT345
To Be Published
|
|
7QC1
 
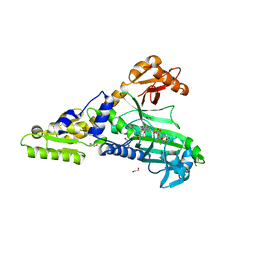 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT436 | | Descriptor: | 1,2-ETHANEDIOL, Proline--tRNA ligase, [(2~{R},3~{S})-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)-2-oxidanylidene-propyl]piperidin-3-yl] ~{N}-[4-[[3-(2,3-dihydro-1~{H}-inden-2-ylcarbamoyl)pyrazin-2-yl]carbamoyl]piperazin-1-yl]sulfonylcarbamate | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT436
To Be Published
|
|
7QC2
 
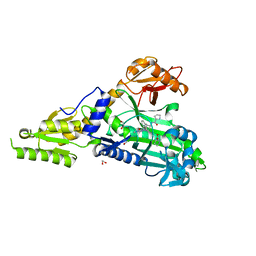 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT334 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT334 and L-Proline
To Be Published
|
|
6I8L
 
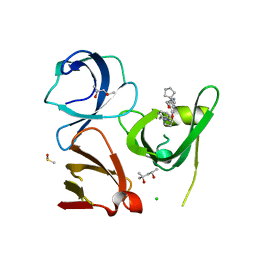 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6QPL
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor MS31 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Johansson, C, Krojer, T, Xiong, Y, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-02-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Potent and Selective Fragment-like Inhibitor of Methyllysine Reader Protein Spindlin 1 (SPIN1).
J.Med.Chem., 62, 2019
|
|
6RBJ
 
 | | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, CHLORIDE ION, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Crystal structure of KDM3B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
6RBI
 
 | | Crystal structure of KDM5B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol | | Descriptor: | 1,2-ETHANEDIOL, 5-(1~{H}-1,2,3,4-tetrazol-5-yl)quinolin-8-ol, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Johansson, C, Newman, J.A, Kawamura, A, Schofield, C.J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-04-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of KDM5B in complex with 5-(1H-tetrazol-5-yl)quinolin-8-ol
To Be Published
|
|
2FLS
 
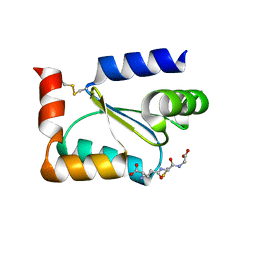 | | Crystal structure of Human Glutaredoxin 2 complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin-2 | | Authors: | Johansson, C, Smee, C, Kavanagh, K.L, Debreczeni, J, von Delft, F, Gileadi, O, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-06 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Human Glutaredoxin 2 complexed with glutathione
To be Published
|
|
2GS3
 
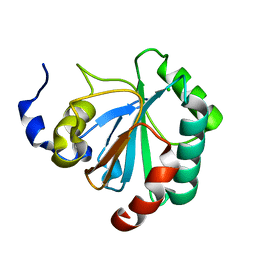 | | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 4(GPX4) | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Johansson, C, Kavanagh, K.L, Rojkova, A, Gileadi, O, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 4(GPX4)
To be Published
|
|
2HT9
 
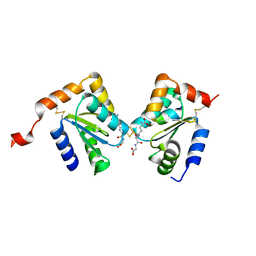 | | The structure of dimeric human glutaredoxin 2 | | Descriptor: | 12-mer peptide, FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, ... | | Authors: | Johansson, C, Smee, C, Kavanagh, K.L, Debreczeni, J, von Delft, F, Gileadi, O, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria
J.Biol.Chem., 282, 2007
|
|
2HE3
 
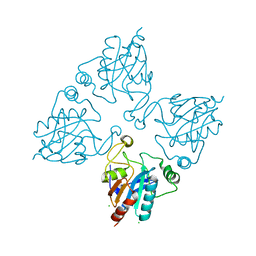 | | Crystal structure of the selenocysteine to cysteine mutant of human glutathionine peroxidase 2 (GPX2) | | Descriptor: | CHLORIDE ION, Glutathione peroxidase 2 | | Authors: | Johansson, C, Kavanagh, K.L, Rojkova, A, Gileadi, O, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the selenocysteine to cysteine mutant of human glutathionine peroxidase 2 (GPX2)
To be Published
|
|
4D6R
 
 | | crystal structure of human JMJD2D in complex with N-OXALYLGLYCINE and bound o-toluenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 4D, N-OXALYLGLYCINE, ... | | Authors: | Krojer, T, Vollmar, M, Bradley, A, Crawley, L, Szykowska, A, Burgess-Brown, N, Gileadi, C, Johansson, C, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystal Structure of Human Jmjd2D in Complex with N-Oxalylglycine and Bound O-Toluenesulfonamide
To be Published
|
|
4UF0
 
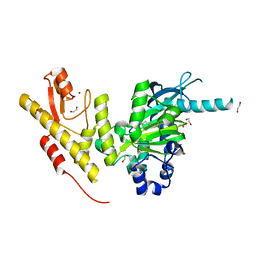 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
4UY4
 
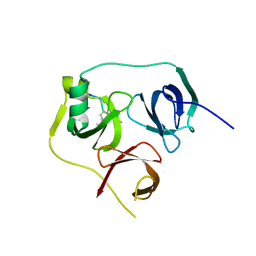 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | Descriptor: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | Authors: | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
2C46
 
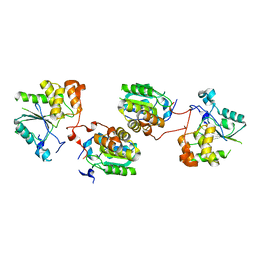 | | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | | Descriptor: | MRNA CAPPING ENZYME | | Authors: | Debreczeni, J, Johansson, C, Longman, E, Gileadi, O, SavitskySmee, P, Smee, C, Bunkoczi, G, Ugochukwu, E, von Delft, F, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-10-15 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Human RNA Guanylyltransferase and 5'-Phosphatase
To be Published
|
|
3BYI
 
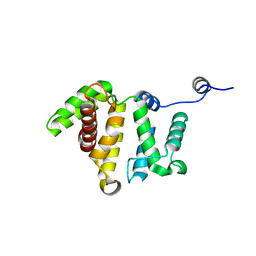 | | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) | | Descriptor: | Rho GTPase activating protein 15 | | Authors: | Shrestha, L, Tickle, J, Elkins, J, Burgess-Brown, N, Johansson, C, Papagrigoriou, E, Kavanagh, K, Pike, A.C.W, Ugochukwu, E, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rho GTPase Activating Protein 15 (ARHGAP15).
To be Published
|
|
5RAE
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001558a | | Descriptor: | 3-[4-(4-hydroxyphenyl)phenyl]propanoic acid, CHLORIDE ION, Lysine-specific demethylase 3B, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
5RAV
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD1B in complex with FM001763a | | Descriptor: | CHLORIDE ION, Lysine-specific demethylase 3B, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of Human JMJD1B screened against the DSPL Fragment Library
To Be Published
|
|
