1EIA
 
 | |
2EIA
 
 | |
7C4Q
 
 | | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), ... | | Authors: | Jin, Z, Park, J.H, Yun, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA
To Be Published
|
|
6AK2
 
 | | Crystal structure of the syntenin PDZ1 domain in complex with the peptide inhibitor KSL-128018 | | Descriptor: | Syntenin-1, peptide inhibitor KSL-128018 | | Authors: | Jin, Z.Y, Park, J.H, Yun, J.H, Haugaard-Kedstrom, L.M, Lee, W.T. | | Deposit date: | 2018-08-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | A High-Affinity Peptide Ligand Targeting Syntenin Inhibits Glioblastoma.
J.Med.Chem., 64, 2021
|
|
7C4P
 
 | | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), ... | | Authors: | Jin, Z, Park, J.H, Yun, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of DBD plasma treated zebrafish TRF2 myb-domain complexed with DNA
To Be Published
|
|
7C4R
 
 | | Crystal structure of hydrogen peroxide treated zebrafish TRF2 complexed with DNA | | Descriptor: | DNA (5'-D(*D*CP*DP*CP*DP*CP*DP*TP*DP*AP*DP*AP*DP*CP*DP*CP*DP*CP*DP*TP*DP*AP*DP*A)-3'), DNA (5'-D(*D*TP*DP*TP*DP*AP*DP*GP*DP*GP*DP*GP*DP*TP*DP*TP*DP*AP*DP*G)-3'), DNA (5'-D(*D*TP*DP*TP*DP*AP*DP*GP*DP*GP*DP*GP*DP*TP*DP*TP*DP*AP*DP*GP*DP*GP*DP*G)-3'), ... | | Authors: | Jin, Z, Park, J.H, Yun, J.H, Park, S.Y, Lee, W. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of hydrogen peroxide treated zebrafish TRF2 myb-domain complexed with DNA
To Be Published
|
|
4NAD
 
 | |
4NAC
 
 | |
8FU3
 
 | | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | | Descriptor: | 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide, Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Yu, X, Abeywickrema, P, Bonneux, B, Behera, I, Jacoby, E, Fung, A, Adhikary, S, Bhaumik, A, Carbajo, R.J, Bruyn, S.D, Miller, R, Patrick, A, Pham, Q, Piassek, M, Verheyen, N, Shareef, A, Sutto-Ortiz, P, Ysebaert, N, Vlijmen, H.V, Jonckers, T.H.M, Herschke, F, McLellan, J.S, Decroly, E, Fearns, R, Grosse, S, Roymans, D, Sharma, S, Rigaux, P, Jin, Z. | | Deposit date: | 2023-01-16 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor.
Commun Biol, 6, 2023
|
|
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
3D2P
 
 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-arginine | | Descriptor: | ARGININE, COENZYME A, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
3D2M
 
 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-glutamate | | Descriptor: | COENZYME A, GLUTAMIC ACID, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
4NEX
 
 | | Structure of the N-acetyltransferase domain of X. fastidiosa NAGS/K | | Descriptor: | Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE, SULFATE ION | | Authors: | Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2013-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6955 Å) | | Cite: | Structures of the N-acetyltransferase domain of Xylella fastidiosaN-acetyl-L-glutamate synthase/kinase with and without a His tag bound to N-acetyl-L-glutamate.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
2R8V
 
 | | Native structure of N-acetylglutamate synthase from Neisseria gonorrhoeae | | Descriptor: | ACETYL COENZYME *A, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-09-11 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
2R98
 
 | | Crystal Structure of N-acetylglutamate synthase (selenoMet substituted) from Neisseria gonorrhoeae | | Descriptor: | ACETYL COENZYME *A, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-09-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
7FCI
 
 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
7VRF
 
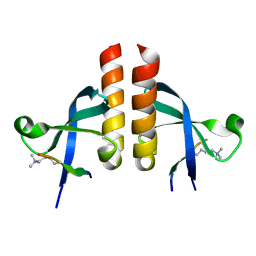 | | Crystal structure of Oxpecker chromodomain in complex with H3K9me3 | | Descriptor: | H3K9me3, Oxpecker | | Authors: | Huang, Y, Jin, Z, Yu, B. | | Deposit date: | 2021-10-22 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the chromodomain of Oxpecker in complex with histone H3 lysine 9 trimethylation reveal a transposon silencing mechanism by heterodimerization.
Biochem.Biophys.Res.Commun., 652, 2023
|
|
6AB9
 
 | | The crystal structure of the relaxed state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven cyanobacterial chloride importer in the rhodopsin family
To Be Published
|
|
6ABA
 
 | | The crystal structure of the photoactivated state of Nonlabens marinus Rhodopsin 3 | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.-H, Ohki, M, Park, J.-H, Jin, Z, Lee, W, Liu, H, Tame, J.R.H, Shibayama, N, Park, S.-Y. | | Deposit date: | 2018-07-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The pumping mechanism of NM-R3, a light-driven marine bacterial chloride importer in the rhodopsin family
To Be Published
|
|
3B8G
 
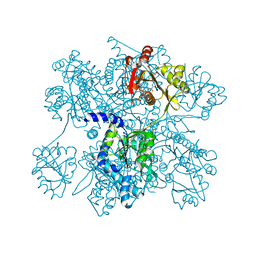 | | Crysta structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and N-acetyl-glutamate | | Descriptor: | COENZYME A, N-ACETYL-L-GLUTAMATE, Putative acetylglutamate synthase | | Authors: | Shi, D, Sagar, V, Jin, Z, Yu, X, Caldovic, L, Morizono, H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2007-11-01 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of N-acetyl-L-glutamate synthase from Neisseria gonorrhoeae provides insights into mechanisms of catalysis and regulation.
J.Biol.Chem., 283, 2008
|
|
4NF1
 
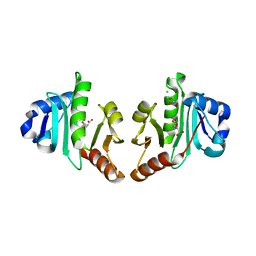 | | Structure of N-acetyltransferase domain of X. fastidiosa NAGS/K without his-tag | | Descriptor: | Acetylglutamate kinase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2013-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3986 Å) | | Cite: | Structures of the N-acetyltransferase domain of Xylella fastidiosaN-acetyl-L-glutamate synthase/kinase with and without a His tag bound to N-acetyl-L-glutamate.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
