8K2W
 
 | | Structure of CXCR3 complexed with antagonist AMG487 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, N-[(1R)-1-[3-(4-ethoxyphenyl)-4-oxidanylidene-pyrido[2,3-d]pyrimidin-2-yl]ethyl]-N-(pyridin-3-ylmethyl)-2-[4-(trifluoromethyloxy)phenyl]ethanamide, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
8K2X
 
 | | CXCR3-DNGi complex activated by CXCL10 | | Descriptor: | C-X-C motif chemokine 10, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487.
Cell Discov, 9, 2023
|
|
6LV0
 
 | |
6IG2
 
 | | Structure of mitochondrial CDP-DAG synthase Tam41 complexed with CTP, delta 74, F240A | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Phosphatidate cytidylyltransferase, mitochondrial | | Authors: | Jiao, H.Z, Yin, Y, Liu, Z.F. | | Deposit date: | 2018-09-23 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structures of the Mitochondrial CDP-DAG Synthase Tam41 Suggest a Potential Lipid Substrate Pathway from Membrane to the Active Site.
Structure, 27, 2019
|
|
6IG4
 
 | | Structure of mitochondrial CDP-DAG synthase Tam41, delta 74 | | Descriptor: | PLATINUM (II) ION, Phosphatidate cytidylyltransferase, mitochondrial, ... | | Authors: | Jiao, H.Z, Yin, Y, Liu, Z.F. | | Deposit date: | 2018-09-23 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Structures of the Mitochondrial CDP-DAG Synthase Tam41 Suggest a Potential Lipid Substrate Pathway from Membrane to the Active Site.
Structure, 27, 2019
|
|
8HNK
 
 | | CXCR3-DNGi complex activated by CXCL11 | | Descriptor: | C-X-C motif chemokine 11, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNL
 
 | | CXCR3-DNGi complex activated by PS372424 | | Descriptor: | (3S)-N-[(2S)-5-carbamimidamido-1-(cyclohexylmethylamino)-1-oxidanylidene-pentan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1H-isoquinoline-3-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNM
 
 | | CXCR3-DNGi complex activated by VUF11222 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | Descriptor: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WBG
 
 | |
8WBH
 
 | |
8WBF
 
 | |
8WBC
 
 | |
8WBB
 
 | |
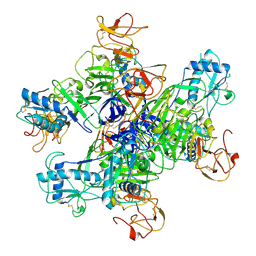
8WBE
 
 | |
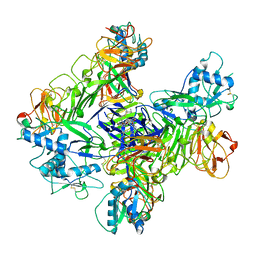
8WBD
 
 | |
6LVF
 
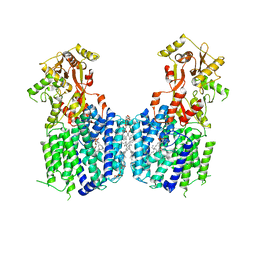 | | Cryo-EM structure of the multiple peptide resistance factor (MprF) loaded with one lysyl-phosphatidylglycerol molecule | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Low pH-inducible protein LpiA, ... | | Authors: | Song, D.F, Jiao, H.Z, Liu, Z.F. | | Deposit date: | 2020-02-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Phospholipid translocation captured in a bifunctional membrane protein MprF.
Nat Commun, 12, 2021
|
|
7DUW
 
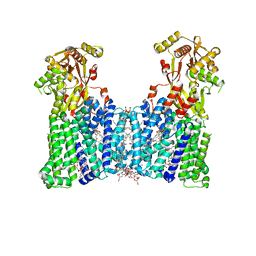 | | Cryo-EM structure of the multiple peptide resistance factor (MprF) loaded with two lysyl-phosphatidylglycerol molecules | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2~{R},3~{S},4~{S},5~{S},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{S},4~{R},5~{R},6~{R})-2-(hydroxymethyl)-6-[2-[[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]oxymethyl]-4-[(1~{R},2~{R},4~{S},5'~{R},6~{R},7~{R},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxy-butoxy]-4,5-bis(oxidanyl)oxan-3-yl]oxy-oxane-3,4,5-triol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Song, D.F, Jiao, H.Z, Liu, Z.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Phospholipid translocation captured in a bifunctional membrane protein MprF.
Nat Commun, 12, 2021
|
|
8XI3
 
 | | Structure of mouse SCMC-14-3-3gama complex | | Descriptor: | 14-3-3 protein gamma, NACHT, LRR and PYD domains-containing protein 5, ... | | Authors: | Chi, P, Han, Z, Jiao, H, Li, J, Wang, X, Hu, H, Deng, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of mouse SCMC-14-3-3gama complex
To Be Published
|
|
