7NL6
 
 | | Crystal Structure of DC-SIGN in complex with a triazole-based glycomimetic ligand | | Descriptor: | CALCIUM ION, DC-SIGN, CRD domain, ... | | Authors: | Jakob, R.P, Cramer, J, Lakkaichi, A, Aliu, B, Cattaneo, I, Klein, S, Jiang, X, Rabbani, S, Schwardt, O, Ernst, B, Maier, T. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sweet Drugs for Bad Bugs: A Glycomimetic Strategy against the DC-SIGN-Mediated Dissemination of SARS-CoV-2.
J.Am.Chem.Soc., 143, 2021
|
|
1Z1D
 
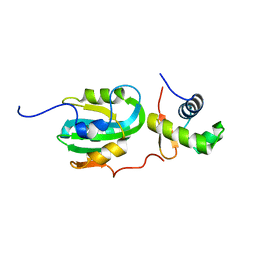 | | Structural Model for the interaction between RPA32 C-terminal domain and SV40 T antigen origin binding domain. | | Descriptor: | Large T antigen, Replication protein A 32 kDa subunit | | Authors: | Arunkumar, A.I, Klimovich, V, Jiang, X, Ott, R.D, Mizoue, L, Fanning, E, Chazin, W.J. | | Deposit date: | 2005-03-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.
Nat.Struct.Mol.Biol., 12, 2005
|
|
5WBI
 
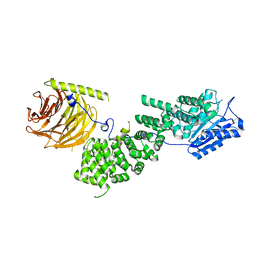 | |
6UT9
 
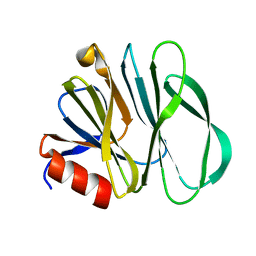 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[4] rotavirus strain BM5265 | | Descriptor: | Outer capsid protein VP4 | | Authors: | Xu, S, Stuckert, M, Burnside, R, McGinnis, K, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5WBJ
 
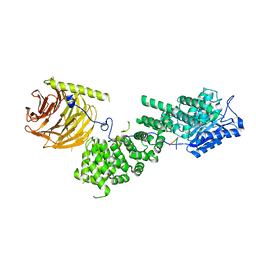 | |
5K0X
 
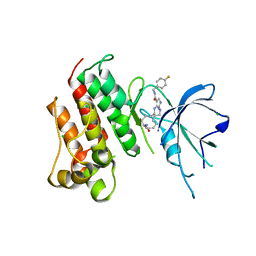 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
6VKX
 
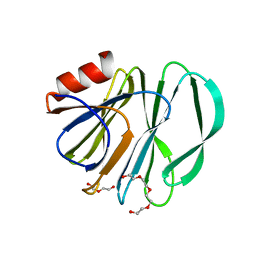 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[8] rotavirus strain BM13851 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Outer capsid protein VP4, TETRAETHYLENE GLYCOL | | Authors: | Xu, S, McGinnis, K.R, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5ERG
 
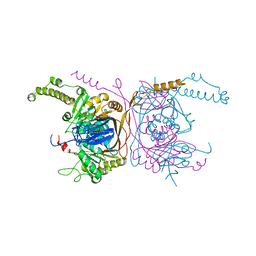 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 in complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5EQJ
 
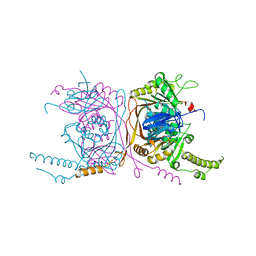 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
5DPM
 
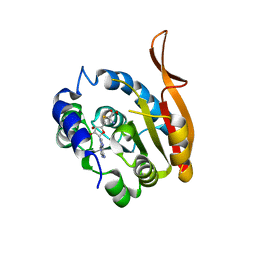 | |
4X5R
 
 | | Crystal structure of FimH in complex with a squaryl-phenyl alpha-D-mannopyranoside derivative | | Descriptor: | 2-chloro-4-{[2-(4-methylpiperazin-1-yl)-3,4-dioxocyclobut-1-en-1-yl]amino}phenyl alpha-D-mannopyranoside, Protein FimH, SULFATE ION | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X5P
 
 | | Crystal structure of FimH in complex with a benzoyl-amidophenyl alpha-D-mannopyranoside | | Descriptor: | 4-{[3-chloro-4-(alpha-D-mannopyranosyloxy)phenyl]carbamoyl}benzoic acid, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.997 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X50
 
 | | Crystal structure of FimH in complex with biphenyl alpha-D-mannopyranoside | | Descriptor: | Protein FimH, biphenyl-4-yl alpha-D-mannopyranoside | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
4X5Q
 
 | | Crystal structure of FimH in complex with 5-nitro-indolinylphenyl alpha-D-mannopyranoside | | Descriptor: | 4-(5-nitro-1H-indol-1-yl)phenyl alpha-D-mannopyranoside, Protein FimH | | Authors: | Preston, R.C, Jakob, R.P, Fiege, B, Zihlmann, P, Rabbani, S, Schwardt, O, Jiang, X, Ernst, B, Maier, T. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Chembiochem, 16, 2015
|
|
6LYY
 
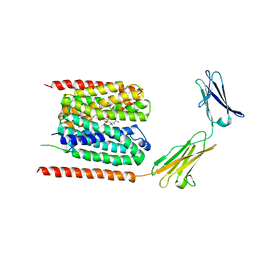 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate AZD3965 in the outward-open conformation. | | Descriptor: | 3-methyl-5-[[(4~{R})-4-methyl-4-oxidanyl-1,2-oxazolidin-2-yl]carbonyl]-6-[[5-methyl-3-(trifluoromethyl)-1~{H}-pyrazol-4-yl]methyl]-1-propan-2-yl-thieno[2,3-d]pyrimidine-2,4-dione, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
7SPT
 
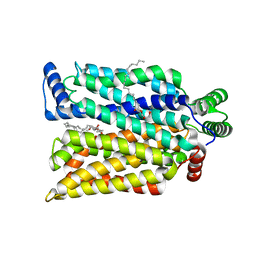 | | Crystal structure of exofacial state human glucose transporter GLUT3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
7SPS
 
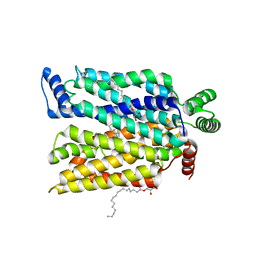 | | Crystal structure of human glucose transporter GLUT3 bound with exofacial inhibitor SA47 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2021-11-03 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for inhibiting human glucose transporters by exofacial inhibitors.
Nat Commun, 13, 2022
|
|
6LZ0
 
 | | Cryo-EM structure of human MCT1 in complex with Basigin-2 in the presence of lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6NIW
 
 | | Crystal structure of P[6] rotavirus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease-sensitive outer capsid protein | | Authors: | Xu, S, Liu, Y, Lakamp, L, Ahmed, L, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
6OAI
 
 | | Crystal structure of P[6] rotavirus vp8* complexed with LNFPI | | Descriptor: | Protease-sensitive outer capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Liu, Y, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
4ZCH
 
 | | Single-chain human APRIL-BAFF-BAFF Heterotrimer | | Descriptor: | TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, Tumor necrosis factor ligand superfamily member 13,Tumor necrosis factor ligand superfamily member 13B,Tumor necrosis factor ligand superfamily member 13B | | Authors: | Lammens, A, Jiang, X, Maskos, K, Schneider, P. | | Deposit date: | 2015-04-16 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Stoichiometry of Heteromeric BAFF and APRIL Cytokines Dictates Their Receptor Binding and Signaling Properties.
J.Biol.Chem., 290, 2015
|
|
5DAF
 
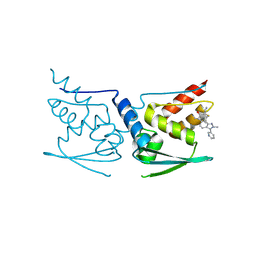 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64063 | | Descriptor: | (5aS,6S,9aS)-7-hydroxy-2,6,9a-trimethyl-3-(pyridin-3-yl)-4,5,5a,6,9,9a-hexahydro-2H-benzo[g]indazole-8-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
5DAD
 
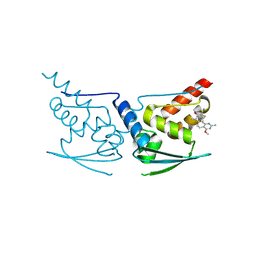 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64014 | | Descriptor: | (6aS,7S,10aS)-8-hydroxy-4-methoxy-2,7,10a-trimethyl-5,6,6a,7,10,10a-hexahydrobenzo[h]quinazoline-9-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
6C9W
 
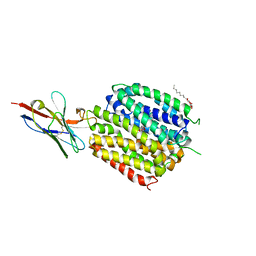 | | Crystal Structure of a ligand bound LacY/Nanobody Complex | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, Nanobody9047, ... | | Authors: | Kumar, H, Finer-Moore, J.S, Jiang, X, Smirnova, I, Kasho, V, Pardon, E, Steyaert, J, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a ligand-bound LacY-Nanobody Complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
