3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
6L95
 
 | | transmembrane-domain of Bax | | Descriptor: | Apoptosis regulator BAX | | Authors: | Bo, O.Y, Fujiao, L. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | transmembrane-domain of Bax
To Be Published
|
|
6LS5
 
 | | Structure of human liver FBPase complexed with covalent allosteric inhibitor | | Descriptor: | 2-(ethyldisulfanyl)-1,3-benzothiazole, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Rongrong, S, Yixiang, X, Shuaishuai, N, Yanliang, R, Jian, L, Jian, W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction.
J.Med.Chem., 63, 2020
|
|
6K0O
 
 | | The crystal structure of human CD163-like homolog SRCR8 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M160 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
6L05
 
 | | Crystal structure of uPA_H99Y in complex with 50F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-6-(1-benzofuran-2-yl)-Ncarbamimidoyl-pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of uPA_H99Y in complex with 50F
To Be Published
|
|
6L04
 
 | | Crystal structure of uPA_H99Y in complex with 31F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of uPA_H99Y in complex with 31F
To Be Published
|
|
8THZ
 
 | | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CBH-7 Heavy chain, ... | | Authors: | Shahid, S, Jiang, L, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2
To Be Published
|
|
1XXE
 
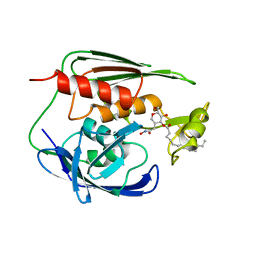 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
7VUX
 
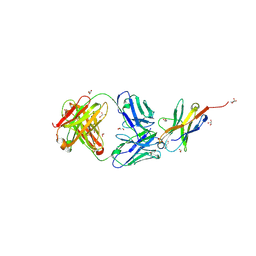 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
7TD5
 
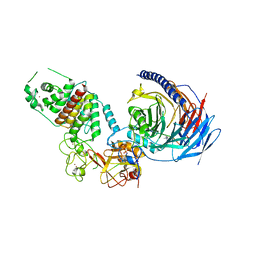 | | Structure of human PRC2-EZH1 containing phosphorylated SUZ12 | | Descriptor: | Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Gong, L, Jiao, L, Liu, X. | | Deposit date: | 2021-12-30 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | CK2-mediated phosphorylation of SUZ12 promotes PRC2 function by stabilizing enzyme active site.
Nat Commun, 13, 2022
|
|
2K4U
 
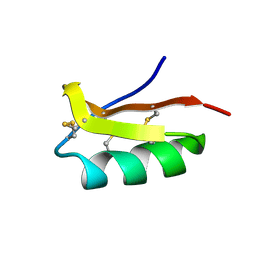 | | Solution structure of the SCORPION TOXIN ADWX-1 | | Descriptor: | Potassium channel toxin alpha-KTx 3.6 | | Authors: | Yin, S.J, Jiang, L, Yi, H, Han, S, Yang, D.W, Liu, M.L, Liu, H, Cao, Z.J, Wu, Y.L, Li, W.X. | | Deposit date: | 2008-06-18 | | Release date: | 2008-12-09 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Different Residues in Channel Turret Determining the Selectivity of ADWX-1 Inhibitor Peptide between Kv1.1 and Kv1.3 Channels
J.Proteome Res., 7, 2008
|
|
4QNP
 
 | | Crystal structure of the 2009 pandemic H1N1 influenza virus neuraminidase with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wan, H.Q, Yang, H, Shore, D.A, Garten, R.J, Couzens, L, Gao, J, Jiang, L.L, Carney, P.J, Villanueva, J, Stevens, J, Eichelberger, M.C. | | Deposit date: | 2014-06-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers.
Nat Commun, 6, 2015
|
|
3P71
 
 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
5Z65
 
 | | Crystal structure of porcine aminopeptidase N ectodomain in functional form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, Y.G, Li, R, Jiang, L.G, Qiao, S.L, Zhang, G.P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization of the interaction between recombinant porcine aminopeptidase N and spike glycoprotein of porcine epidemic diarrhea virus.
Int. J. Biol. Macromol., 117, 2018
|
|
5F8X
 
 | | The crystal structure of human plasma kallikrein in complex with its peptide inhibitor pkalin-3 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-TRP-CYS, Plasma kallikrein, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure Of Human Plasma Kallikrein In Complex With Its Peptide Inhibitor Pkalin-3
To Be Published
|
|
6A8G
 
 | | The crystal structure of muPAin-1-IG in complex with muPA-SPD at pH8.5 | | Descriptor: | PHOSPHATE ION, Urokinase-type plasminogen activator chain B, muPAin-1-IG | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-08 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
6XVD
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3F) in pH7.4 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1-W3F | | Authors: | Xue, G.P, Xie, X, Zhou, Y, Yuan, C, Huang, M.D, Jiang, L.G. | | Deposit date: | 2020-01-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
8ITD
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(4-chloranyl-2-oxidanyl-phenyl)-2-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8IT8
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(4-chlorophenyl)-2-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8IT7
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(4-chlorophenyl)-2-oxidanyl-6-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8ITC
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(4-chlorophenyl)-3-[(4-phenylphenyl)sulfonylamino]phthalic acid, CHLORIDE ION, ... | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
8IT4
 
 | | Phosphoglycerate mutase 1 complexed with a covalent inhibitor | | Descriptor: | 3-[[5-(4-chlorophenyl)-2-methoxycarbonyl-thiophen-3-yl]sulfamoyl]benzenesulfonic acid, CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a covalent inhibitor
To Be Published
|
|
8IT6
 
 | | Phosphoglycerate mutase 1 complexed with a compound | | Descriptor: | 4-[4-chloranyl-2-(2-hydroxy-2-oxoethyloxy)phenyl]-2-[(4-phenylphenyl)sulfonylamino]benzoic acid, CHLORIDE ION, Phosphoglycerate mutase 1 | | Authors: | Zhou, L, Jiang, L.L. | | Deposit date: | 2023-03-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphoglycerate mutase 1 complexed with a compound
To Be Published
|
|
