2SGA
 
 | |
2PSG
 
 | |
4SGA
 
 | |
2SGQ
 
 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
1SGT
 
 | |
3CAG
 
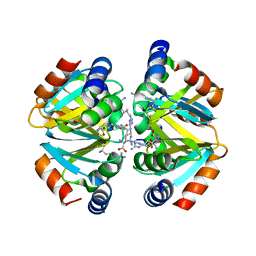 | | Crystal structure of the oligomerization domain hexamer of the arginine repressor protein from Mycobacterium tuberculosis in complex with 9 arginines. | | Descriptor: | ARGININE, Arginine repressor | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-02-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
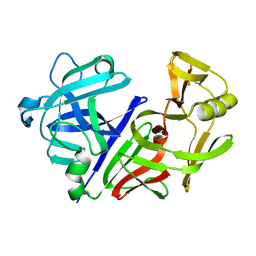
3APP
 
 | |
2SGP
 
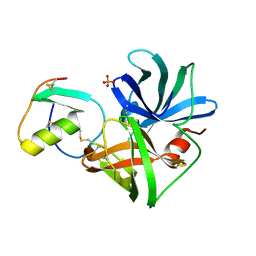 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2001-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
3SGA
 
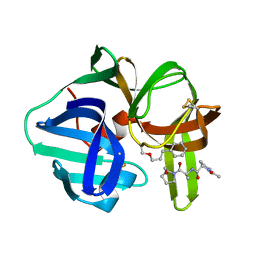 | |
6AQ7
 
 | | Structure of POM6 FAB fragment complexed with mouse PrPc | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GLYCEROL, Major prion protein, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of POM6 Fab and mouse prion protein complex identifies key regions for prions conformational conversion.
FEBS J., 285, 2018
|
|
2SEC
 
 | |
3SGB
 
 | | STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | PROTEINASE B (SGPB), TURKEY OVOMUCOID INHIBITOR (OMTKY3) | | Authors: | Read, R.J, Fujinaga, M, Sielecki, A.R, James, M.N.G. | | Deposit date: | 1983-01-21 | | Release date: | 1983-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the complex of Streptomyces griseus protease B and the third domain of the turkey ovomucoid inhibitor at 1.8-A resolution.
Biochemistry, 22, 1983
|
|
1APW
 
 | |
5TNC
 
 | |
1APT
 
 | |
1APU
 
 | |
5SGA
 
 | |
1TON
 
 | |
1U32
 
 | | Crystal structure of a Protein Phosphatase-1: Calcineurin Hybrid Bound to Okadaic Acid | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Perreault, K.R, Cherney, M.M, Luu, H.A, James, M.N.G, Holmes, C.F.B. | | Deposit date: | 2004-07-20 | | Release date: | 2004-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Mutagenesis of a Protein Phosphatase-1:Calcineurin Hybrid Elucidate the Role of the {beta}12-{beta}13 Loop in Inhibitor Binding
J.Biol.Chem., 279, 2004
|
|
1TJN
 
 | | Crystal structure of hypothetical protein af0721 from Archaeoglobus fulgidus | | Descriptor: | Sirohydrochlorin cobaltochelatase | | Authors: | Yin, J, Xu, X.L, Cuff, M, Walker, J.R, Edwards, A, Savchenko, A, James, M.N.G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of af0721: a hypothetical protein bearing sequence similarity with class II chelatases in cobalamin synthesis
To be Published
|
|
3NAG
 
 | | Crystal structure of the phosphoribosylpyrophosphate (PRPP) synthetase from Thermoplasma Volcanium in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribose-phosphate pyrophosphokinase, ... | | Authors: | Cherney, M.M, Cherney, L.T, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
1APV
 
 | |
2GT7
 
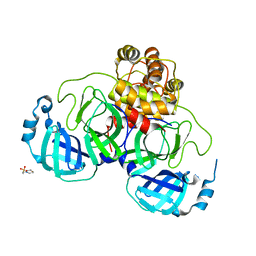 | | Crystal structure of SARS coronavirus main peptidase at pH 6.0 in the space group P21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2GT8
 
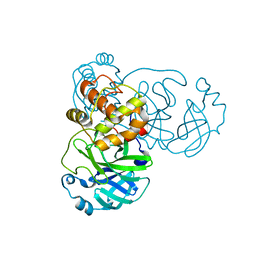 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) in the space group P43212 | | Descriptor: | 3C-like proteinase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N.G. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2REN
 
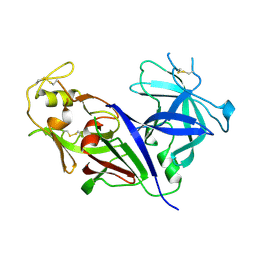 | | STRUCTURE OF RECOMBINANT HUMAN RENIN, A TARGET FOR CARDIOVASCULAR-ACTIVE DRUGS, AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RENIN | | Authors: | Sielecki, A.R, James, M.N.G. | | Deposit date: | 1992-02-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of recombinant human renin, a target for cardiovascular-active drugs, at 2.5 A resolution.
Science, 243, 1989
|
|
