7SP2
 
 | | Structure of PLS A-domain (residues 391-656; 513-518 deletion mutant) from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Plasmin Sensitive Protein Pls | | Authors: | Clark, L, Whelan, F, Atkin, K.E, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-11-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of PLS A-domain (residues 391-65) from Staphylococcus aureus
Not Published
|
|
1R9D
 
 | | Glycerol bound form of the B12-independent glycerol dehydratase from Clostridium butyricum | | Descriptor: | GLYCEROL, glycerol dehydratase | | Authors: | Lanzilotta, W.N, O'Brien, J.R, Raynaud, C, Soucaille, P. | | Deposit date: | 2003-10-28 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization.
Biochemistry, 43, 2004
|
|
1RGZ
 
 | | Enterobacter cloacae GC1 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | GLYCEROL, class C beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S3Z
 
 | |
1U8E
 
 | | HUMAN DIPEPTIDYL PEPTIDASE IV/CD26 MUTANT Y547F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bjelke, J.R, Christensen, J, Branner, S, Wagtmann, N, Olsen, C, Kanstrup, A.B, Rasmussen, H.B. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 547 constitutes an essential part of the catalytic mechanism of dipeptidyl peptidase IV
J.Biol.Chem., 279, 2004
|
|
3S1J
 
 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3SCU
 
 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with Cellopentaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
1S3V
 
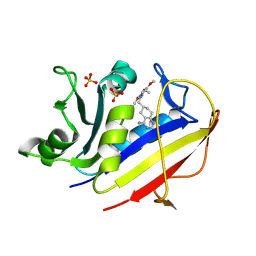 | | Structure Determination of Tetrahydroquinazoline Antifolates in Complex with Human and Pneumocystis carinii Dihydrofolate Reductase: Correlations of Enzyme Selectivity and Stereochemistry | | Descriptor: | (2R,6S)-6-{[methyl(3,4,5-trimethoxyphenyl)amino]methyl}-1,2,5,6,7,8-hexahydroquinazoline-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A, Queener, S.F. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of tetrahydroquinazoline antifolates in complex with human and Pneumocystis carinii dihydrofolate reductase: correlations between enzyme selectivity and stereochemistry.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3SCO
 
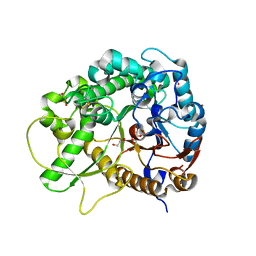 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with alpha-Glucosyl Fluoride | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLYCEROL, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3SCV
 
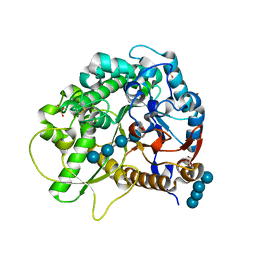 | | Crystal Structure of Rice BGlu1 E386G/S334A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
1S3W
 
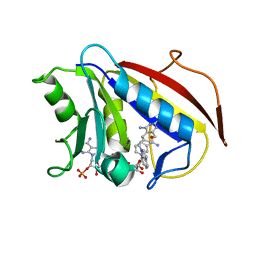 | | Structure Determination of Tetrahydroquinazoline Antifoaltes in Complex with Human and Pneumocystis carinii Dihydrofolate Reductase: Correlations of Enzyme Selectivity and Stereochemistry | | Descriptor: | 6-(OCTAHYDRO-1H-INDOL-1-YLMETHYL)DECAHYDROQUINAZOLINE-2,4-DIAMINE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A, Queener, S.F. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination of tetrahydroquinazoline antifolates in complex with human and Pneumocystis carinii dihydrofolate reductase: correlations between enzyme selectivity and stereochemistry.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3S4Y
 
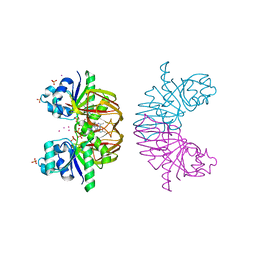 | | Crystal structure of human thiamin pyrophosphokinase 1 | | Descriptor: | CALCIUM ION, SULFATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Shen, L, Tempel, W, Tong, Y, Li, Y, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human thiamin pyrophosphokinase 1
to be published
|
|
7TN0
 
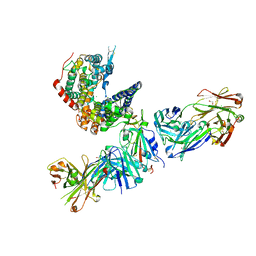 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
3SZM
 
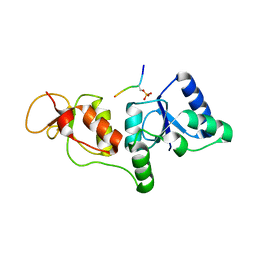 | |
3SVM
 
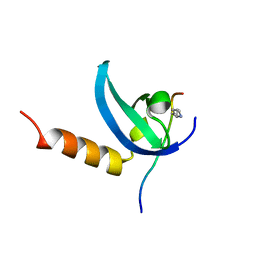 | | Human MPP8 - human DNMT3AK47me2 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
1S05
 
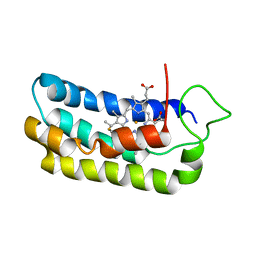 | | NMR-validated structural model for oxidized R.palustris cytochrome c556 | | Descriptor: | Cytochrome c-556, HEME C | | Authors: | Bertini, I, Faraone-Mennella, J, Gray, H.B, Luchinat, C, Parigi, G, Winkler, J.R. | | Deposit date: | 2003-12-30 | | Release date: | 2004-01-20 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | NMR-validated structural model for oxidized Rhodopseudomonas palustris cytochrome c(556).
J.Biol.Inorg.Chem., 9, 2004
|
|
1SGO
 
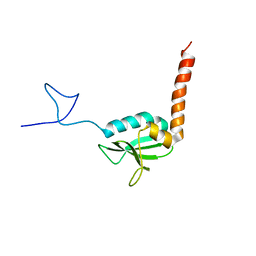 | | NMR Structure of the human C14orf129 gene product, HSPC210. Northeast Structural Genomics target HR969. | | Descriptor: | Protein C14orf129 | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shih, L.-Y, Ma, L.-C, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the human C14orf129 gene product, HSPC210. Northeast Structural Genomics target HR969.
To be Published
|
|
3RHU
 
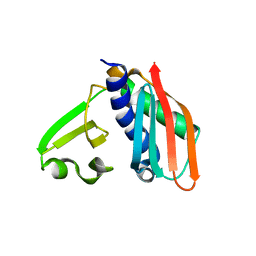 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | SC_1wnu | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
7RR9
 
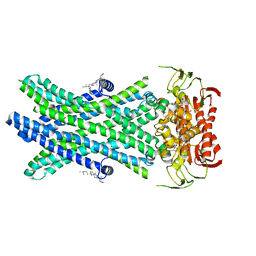 | | Cryo-EM Structure of Nanodisc reconstituted ABCD1 in nucleotide bound outward open conformation | | Descriptor: | ATP-binding cassette sub-family D member 1, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human peroxisomal fatty acid transporter ABCD1 in a lipid environment
Commun Biol, 5, 2022
|
|
7RRA
 
 | |
1SQR
 
 | | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus. Northeast Structural Genomics Consortium Target PfR48. | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L.C, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus: Northeast Structural Genomics Consortium Target PfR48
To be Published
|
|
1T2F
 
 | | Human B lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | | Descriptor: | 4-HYDROXY-1,2,5-OXADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase B chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
3SCW
 
 | | Crystal Structure of Rice BGlu1 E386G/Y341A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
1T64
 
 | | Crystal Structure of human HDAC8 complexed with Trichostatin A | | Descriptor: | CALCIUM ION, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
