3FSS
 
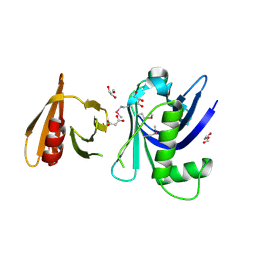 | | Structure of the tandem PH domains of Rtt106 | | Descriptor: | GLYCEROL, Histone chaperone RTT106, MALONIC ACID | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2009-01-11 | | Release date: | 2009-12-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
6OD4
 
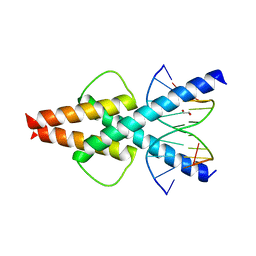 | | Human TCF4 C-terminal bHLH domain in Complex with 11-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*CP*GP*TP*GP*TP*A)-3'), Transcription factor 4 | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
2PI4
 
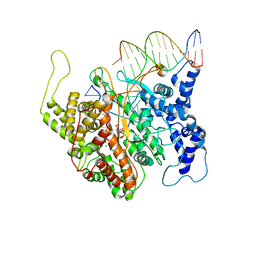 | | T7RNAP complexed with a phi10 protein and initiating GTPs. | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*CP*CP*TP*AP*TP*AP*GP*TP*GP*AP*GP*TP*CP*GP*TP*AP*TP*TP*A)-3', 5'-D(*TP*AP*AP*TP*AP*CP*GP*AP*CP*TP*CP*AP*CP*T)-3', ... | | Authors: | Kennedy, W.P, Momand, J.R, Yin, Y.W. | | Deposit date: | 2007-04-12 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for de novo RNA synthesis and initiating nucleotide specificity by t7 RNA polymerase.
J.Mol.Biol., 370, 2007
|
|
4ZPW
 
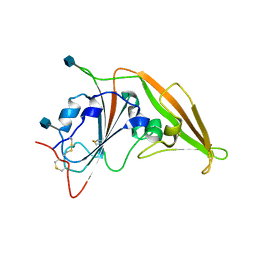 | | Structure of unbound MERS-CoV spike receptor-binding domain (England1 strain). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Spike glycoprotein | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
4A31
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 6-{[2-(4-METHYLPIPERAZIN-1-YL)ETHYL]AMINO}-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)PYRIDINE-3-SULFONAMIDE, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
8BCQ
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (native crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION | | Authors: | Benas, P, Jaramillo Ponce, J.R, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
4A32
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 3,5-DICHLORO-3'-[(DIETHYLAMINO)METHYL]-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BIPHENYL-4-SULFONAMIDE, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
3G93
 
 | | Single ligand occupancy crystal structure of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole | | Descriptor: | 1-(biphenyl-4-ylmethyl)-1H-imidazole, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Sun, L, Maekawa, K, Halpert, J.R, Stout, C.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning.
Biochemistry, 48, 2009
|
|
4ZJH
 
 | | Crystal structure of native alpha-2-macroglobulin from Escherichia coli spanning domains NIE-MG1. | | Descriptor: | ACETATE ION, GLYCEROL, alpha-2-Macroglobulin | | Authors: | Garcia-Ferrer, I, Arede, P, Gomez-Blanco, J, Luque, D, Duquerroy, S, Caston, J.R, Goulas, T, Gomis-Ruth, X.F. | | Deposit date: | 2015-04-29 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional insights into Escherichia coli alpha 2-macroglobulin endopeptidase snap-trap inhibition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2ZD8
 
 | | SHV-1 class A beta-lactamase complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Nukaga, M, Knox, J.R, Hujer, A, Bonomo, R.A. | | Deposit date: | 2007-11-20 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Inhibition of class A beta-lactamases by carbapenems: crystallographic observation of two conformations of meropenem in SHV-1.
J.Am.Chem.Soc., 130, 2008
|
|
8BVO
 
 | |
8BXR
 
 | |
8BW6
 
 | | Titin FnIII-domain I110 (I/A6) from the MIR region | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mayans, O, Fleming, J.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Immunological and Structural Characterization of Titin Main Immunogenic Region; I110 Domain Is the Target of Titin Antibodies in Myasthenia Gravis.
Biomedicines, 11, 2023
|
|
4ZPT
 
 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV
Nat Commun, 6, 2015
|
|
6OAB
 
 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
3G1N
 
 | | Catalytic domain of the human E3 ubiquitin-protein ligase HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, SODIUM ION | | Authors: | Walker, J.R, Qiu, L, Li, Y, Davis, T, Tempel, W, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hect Domain of Human HUWE1/MULE
To be Published
|
|
3FY3
 
 | | Crystal structure of truncated hemolysin A from P. mirabilis | | Descriptor: | Hemolysin | | Authors: | Weaver, T.M, Thompson, J.R, Bailey, L.J, Wawrzyn, G.T, Hocking, J.M, Howard, D.R. | | Deposit date: | 2009-01-21 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional studies of truncated hemolysin A from Proteus mirabilis.
J.Biol.Chem., 284, 2009
|
|
6OHI
 
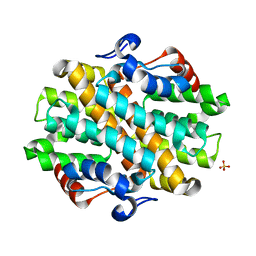 | | Crystal Structure of the Debrominase Bmp8 (Apo) | | Descriptor: | Debrominase Bmp8, SULFATE ION | | Authors: | Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bacterial Tetrabromopyrrole Debrominase Shares a Reductive Dehalogenation Strategy with Human Thyroid Deiodinase.
Biochemistry, 58, 2019
|
|
2Q2G
 
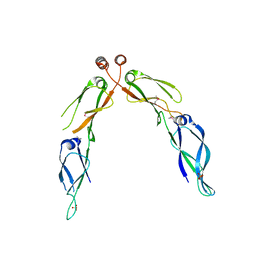 | | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800 | | Descriptor: | Heat shock 40 kDa protein, putative (fragment), SULFATE ION | | Authors: | Wernimont, A.K, Lew, J, Lin, L, Hassanali, A, Kozieradzki, I, Wasney, G, Vedadi, M, Walker, J.R, Zhao, Y, Schapira, M, Bochkarev, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dimerization domain of HSP40 from Cryptosporidium parvum, cgd2_1800.
To be Published
|
|
2Q20
 
 | |
3GJ0
 
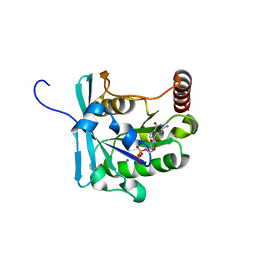 | | Crystal structure of human RanGDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Partridge, J.R, Schwartz, T.U. | | Deposit date: | 2009-03-07 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic and Biochemical Analysis of the Ran-binding Zinc Finger Domain.
J.Mol.Biol., 391, 2009
|
|
3GJ6
 
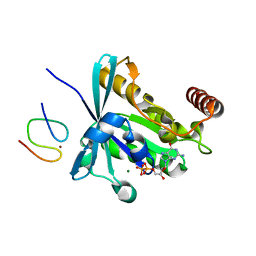 | |
8BS7
 
 | |
4B9H
 
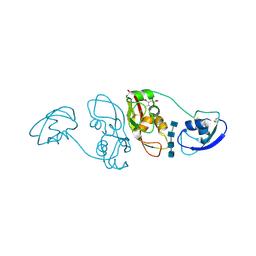 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
