8CD6
 
 | | structure of HEX-1 (cyto V2) from N. crassa grown in living insect cells, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
5UGR
 
 | | Malyl-CoA lyase from Methylobacterium extorquens | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Malyl-CoA lyase/beta-methylmalyl-CoA lyase, ... | | Authors: | Gonzalez, J.M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Methylobacterium extorquens malyl-CoA lyase: CoA-substrate binding correlates with domain shift.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5U9S
 
 | | Ocellatin-F1, solution structure in TFE by NMR spectroscopy | | Descriptor: | Ocellatin-F1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-F1
To Be Published
|
|
5U9Y
 
 | | Ocellatin-F1 | | Descriptor: | Ocellatin-K1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-01 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
2IO6
 
 | | Wee1 kinase complexed with inhibitor PD330961 | | Descriptor: | 9-HYDROXY-6-(3-HYDROXYPROPYL)-4-(2-METHOXYPHENYL)PYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
5TC3
 
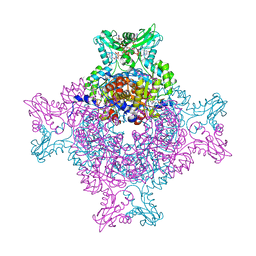 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP and GDP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
2IN6
 
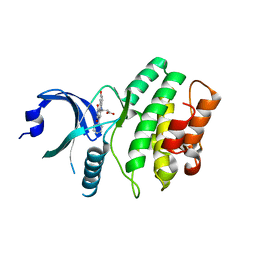 | | Wee1 kinase complex with inhibitor PD311839 | | Descriptor: | 3-(9-HYDROXY-1,3-DIOXO-4-PHENYL-2,3-DIHYDROPYRROLO[3,4-C]CARBAZOL-6(1H)-YL)PROPANOIC ACID, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
8D5C
 
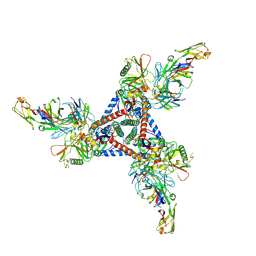 | | anti-HIV-1 gp120-sCD4 complex antibody CG10 Fab in complex with B41-sCD4 | | Descriptor: | CG10 Fab heavy chain, CG10 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | Yang, Z, Bjorkman, P.J, Gershoni, J.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-11-16 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody Recognition of CD4-Induced Open HIV-1 Env Trimers.
J.Virol., 96, 2022
|
|
5T03
 
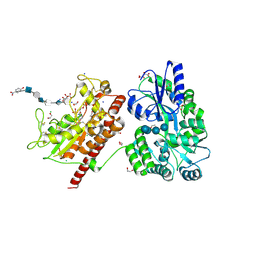 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5TWS
 
 | | Post-catalytic complex of human Polymerase Mu (H329A) with newly incorporated UTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2016-11-14 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural accommodation of ribonucleotide incorporation by the DNA repair enzyme polymerase Mu.
Nucleic Acids Res., 45, 2017
|
|
8DKB
 
 | |
5UB5
 
 | | human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus notch homolog protein 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
8DBE
 
 | | Human PRPS1 with ADP; Hexamer | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBF
 
 | | Human PRPS1 with ADP; Filament Interface | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5T0A
 
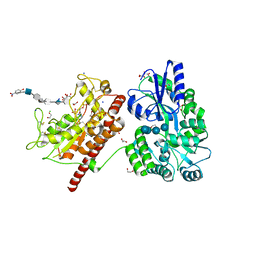 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
8DBO
 
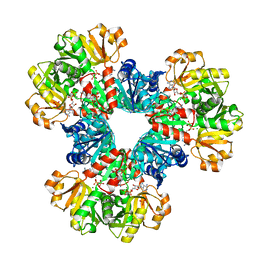 | |
8DBL
 
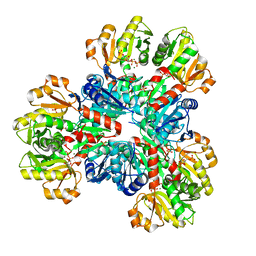 | | Human PRPS1 with Phosphate and PRPP; Hexamer | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBG
 
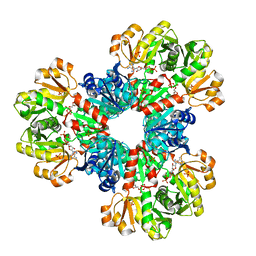 | | Human PRPS1 with Phosphate and ATP; Hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBN
 
 | | Human PRPS1-E307A engineered mutation with Phosphate, ATP, and R5P; Hexamer | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBJ
 
 | | Human PRPS1 with Phosphate, ATP, and R5P; Filament Interface | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBK
 
 | | Human PRPS1 with Phosphate, ATP, and R5P; Hexamer with resolved catalytic loops | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBC
 
 | | Human PRPS1 with Phosphate; Hexamer | | Descriptor: | PHOSPHATE ION, Ribose-phosphate pyrophosphokinase 1 | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBM
 
 | |
8DBD
 
 | |
8DBI
 
 | | Human PRPS1 with Phosphate, ATP, and R5P; Hexamer | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
