5IXA
 
 | |
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
5J34
 
 | | Isopropylmalate dehydrogenase K232M mutant | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
5J04
 
 | |
1FZR
 
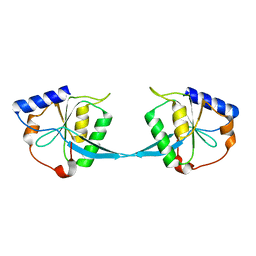 | | CRYSTAL STRUCTURE OF BACTERIOPHAGE T7 ENDONUCLEASE I | | Descriptor: | ENDONUCLEASE I | | Authors: | Hadden, J.M, Convery, M.A, Declais, A.C, Lilley, D.M.J, Phillips, S.E.V. | | Deposit date: | 2000-10-04 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I.
Nat.Struct.Biol., 8, 2001
|
|
1FVT
 
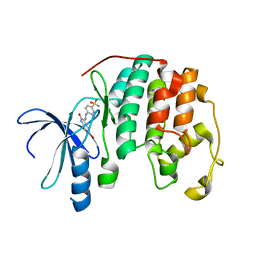 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1G84
 
 | | THE SOLUTION STRUCTURE OF THE C EPSILON2 DOMAIN FROM IGE | | Descriptor: | IMMUNOGLOBULIN E | | Authors: | McDonnell, J.M, Cowburn, D, Gould, H.J, Sutton, B.J, Calvert, R, Beavil, R.E, Beavil, A.J, Henry, A.J. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The structure of the IgE Cepsilon2 domain and its role in stabilizing the complex with its high-affinity receptor FcepsilonRIalpha.
Nat.Struct.Biol., 8, 2001
|
|
1FYY
 
 | | HPRT GENE MUTATION HOTSPOT WITH A BPDE2(10R) ADDUCT | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*TP*GP*CP*CP*CP*TP*TP*GP*AP*CP*TP*A)-3', HPRT DNA WITH BENZO[A]PYRENE-ADDUCTED DA7 | | Authors: | Volk, D.E, Rice, J.S, Luxon, B.A, Yeh, H.J.C, Liang, C, Xie, G, Sayer, J.M, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2000-10-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex.
Biochemistry, 39, 2000
|
|
1G83
 
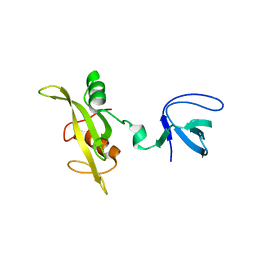 | | CRYSTAL STRUCTURE OF FYN SH3-SH2 | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE FYN | | Authors: | Arold, S.T, Ulmer, T.S, Mulhern, T.D, Werner, J.M, Ladbury, J.E, Campbell, I.D, Noble, M.E.M. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The role of the Src homology 3-Src homology 2 interface in the regulation of Src kinases.
J.Biol.Chem., 276, 2001
|
|
5J1G
 
 | |
1G1Z
 
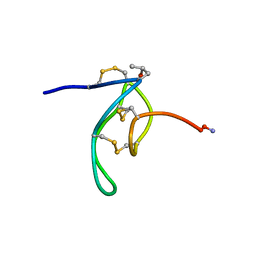 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels, LEU12-PRO13 Cis isomer | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Le Gall, F, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-16 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1G4D
 
 | |
6B3U
 
 | | Solution Structure of HIV-1 GP41 Transmembrane Domain in Bicelles | | Descriptor: | HIV-1 GP41 Transmembrane Domain | | Authors: | Chiliveri, S.C, Louis, J.M, Ghirlando, R, Baber, J.L, Bax, A. | | Deposit date: | 2017-09-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tilted, Uninterrupted, Monomeric HIV-1 gp41 Transmembrane Helix from Residual Dipolar Couplings.
J. Am. Chem. Soc., 140, 2018
|
|
5ISH
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0765 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0765
To Be Published
|
|
5ISE
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0649 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with SAH at 1.8 Angstroms resolution
To Be Published
|
|
1FXH
 
 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
6BU6
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor | | Descriptor: | 4,4'-(2-aminopyridine-3,5-diyl)bis(2,6-difluorophenol), CHLORIDE ION, GLYCEROL, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Santiago, A.S, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a bis-difluorophenol-aminopyridine inhibitor
To Be Published
|
|
1FUU
 
 | | YEAST INITIATION FACTOR 4A | | Descriptor: | YEAST INITIATION FACTOR 4A | | Authors: | Caruthers, J.M, Johnson, E.R, McKay, D.B. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6V3W
 
 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment with four point mutations in complex with RBN-2397 | | Descriptor: | 5-{[(2S)-1-(3-oxo-3-{4-[5-(trifluoromethyl)pyrimidin-2-yl]piperazin-1-yl}propoxy)propan-2-yl]amino}-4-(trifluoromethyl)pyridazin-3(2H)-one, CHLORIDE ION, Protein mono-ADP-ribosyltransferase PARP12 | | Authors: | Swinger, K.K, Gozgit, J.M, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity.
Cancer Cell, 39, 2021
|
|
1FXV
 
 | | PENICILLIN ACYLASE MUTANT IMPAIRED IN CATALYSIS WITH PENICILLIN G IN THE ACTIVE SITE | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE, PENICILLIN G | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-27 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
6BNZ
 
 | | Crystal structure of E144Q-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
5JRI
 
 | | Structure of an oxidoreductase SeMet-labelled from Synechocystis sp. PCC6803 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase, ... | | Authors: | Buey, R.M, de Pereda, J.M, Balsera, M. | | Deposit date: | 2016-05-06 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Unprecedented pathway of reducing equivalents in a diflavin-linked disulfide oxidoreductase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5K8W
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor U2 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U2
To Be Published
|
|
1G1P
 
 | | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus that Selectively Acts on Vertebrate Neuronal Na+ Channels | | Descriptor: | CONOTOXIN EVIA | | Authors: | Volpon, L, Lamthanh, H, Barbier, J, Gilles, N, Molgo, J, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-13 | | Release date: | 2000-11-01 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of delta-Conotoxin EVIA from Conus ermineus That Selectively Acts on Vertebrate Neuronal Na+ Channels.
J.Biol.Chem., 279, 2004
|
|
1GH7
 
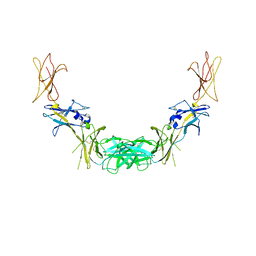 | | CRYSTAL STRUCTURE OF THE COMPLETE EXTRACELLULAR DOMAIN OF THE BETA-COMMON RECEPTOR OF IL-3, IL-5, AND GM-CSF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOKINE RECEPTOR COMMON BETA CHAIN, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carr, P.D, Gustin, S.E, Church, A.P, Murphy, J.M, Ford, S.C, Mann, D.A, Woltring, D.M, Walker, I, Ollis, D.L, Young, I.G. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration.
Cell(Cambridge,Mass.), 104, 2001
|
|
