7SJJ
 
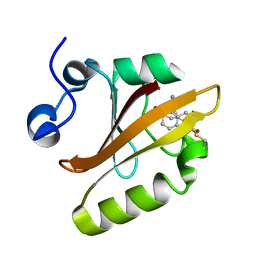 | | Crystal structure of photoactive yellow protein (PYP); F96oCNF construct | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Weaver, J.B, Kirsh, J.M, Boxer, S.G. | | 登録日 | 2021-10-17 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments.
J.Am.Chem.Soc., 144, 2022
|
|
7S6H
 
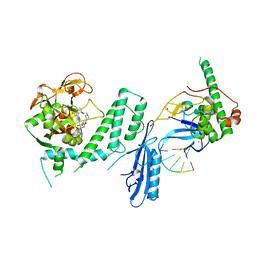 | | Human PARP1 deltaV687-E688 bound to NAD+ analog EB-47 and to a DNA double strand break. | | 分子名称: | 1,2-ETHANEDIOL, 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, DNA (5'-D(*CP*GP*AP*CP*G)-3'), ... | | 著者 | Rouleau-Turcotte, E, Pascal, J.M. | | 登録日 | 2021-09-14 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S81
 
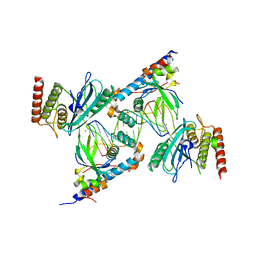 | | Structure of human PARP1 domains (Zn1, Zn3, WGR, HD) bound to a DNA double strand break. | | 分子名称: | DNA (5'-D(*AP*TP*GP*CP*GP*GP*CP*CP*GP*CP*AP*T)-3'), Poly [ADP-ribose] polymerase 1, ZINC ION | | 著者 | Rouleau-Turcotte, E, Pascal, J.M. | | 登録日 | 2021-09-17 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7S6M
 
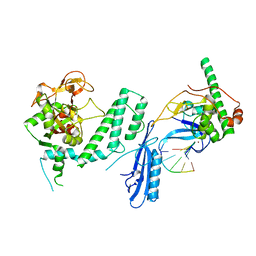 | |
7S68
 
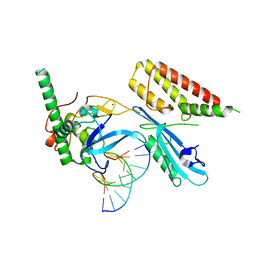 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | 分子名称: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | 著者 | Rouleau-Turcotte, E, Pascal, J.M. | | 登録日 | 2021-09-13 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7RZI
 
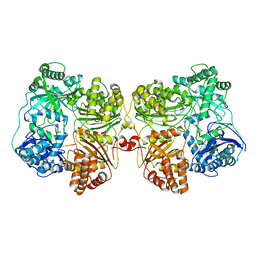 | | Insulin Degrading Enzyme pC/pC | | 分子名称: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
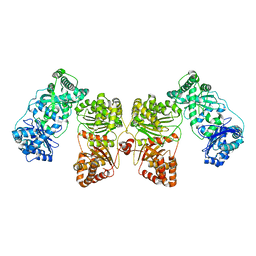 | | Insulin Degrading Enzyme O/O | | 分子名称: | Cysteine-free Insulin-degrading enzyme | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZE
 
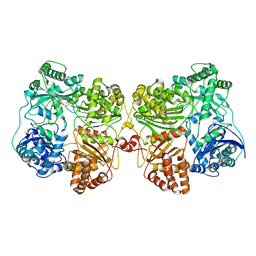 | | Insulin Degrading Enzyme pO/pC | | 分子名称: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
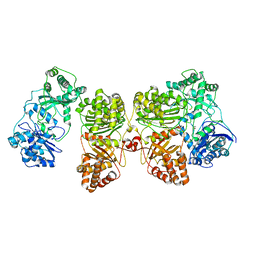 | | Insulin Degrading Enzyme O/pC | | 分子名称: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZG
 
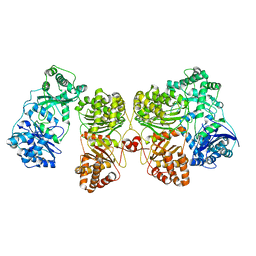 | | Insulin Degrading Enzyme O/pO | | 分子名称: | Cysteine-free Insulin-degrading enzyme | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7S51
 
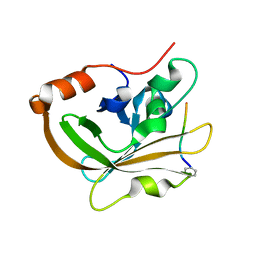 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATA peptide | | 分子名称: | LEU-PRO-ALA-THR-ALA, Sortase | | 著者 | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | 登録日 | 2021-09-09 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
7S4O
 
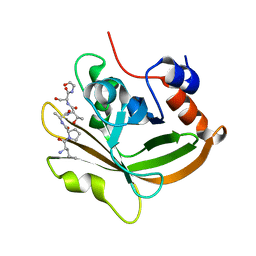 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATS peptide | | 分子名称: | LEU-PRO-ALA-THR-SER-GLY, Sortase | | 著者 | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | 登録日 | 2021-09-09 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.396 Å) | | 主引用文献 | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
5XM3
 
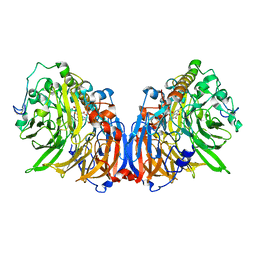 | | Crystal Structure of Methanol dehydrogenase from Methylophaga aminisulfidivorans | | 分子名称: | Glucose dehydrogenase, MAGNESIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | 著者 | Cao, T.P, Choi, J.M, Lee, S.H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | The crystal structure of methanol dehydrogenase, a quinoprotein from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT
J. Microbiol., 56, 2018
|
|
5XWB
 
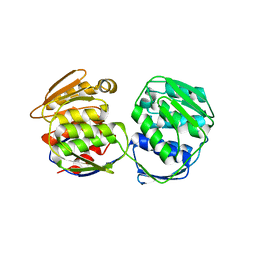 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | 分子名称: | 3-phosphoshikimate 1-carboxyvinyltransferase | | 著者 | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | 登録日 | 2017-06-29 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5WMI
 
 | |
5WP5
 
 | |
5WML
 
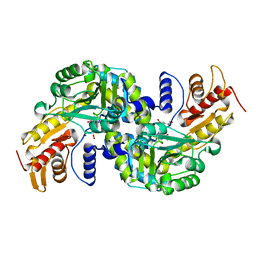 | | Arabidopsis thaliana Prephenate Aminotransferase mutant- K306A | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, GLUTAMIC ACID | | 著者 | Jez, J.M, Holland, C.K. | | 登録日 | 2017-07-29 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
5WP4
 
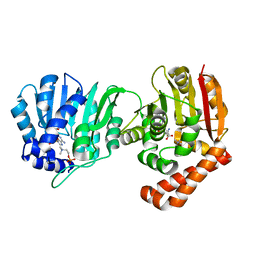 | |
5XMH
 
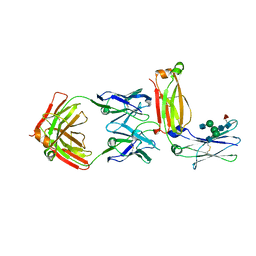 | | Crystal structure of an IgM rheumatoid factor YES8c in complex with IgG1 Fc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, ... | | 著者 | Shiroishi, M, Shimokawa, K, Lee, J.M, Kusakabe, M, Ueda, T. | | 登録日 | 2017-05-15 | | 公開日 | 2018-03-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-function analyses of a stereotypic rheumatoid factor unravel the structural basis for germline-encoded antibody autoreactivity.
J. Biol. Chem., 293, 2018
|
|
5WHF
 
 | |
5WHX
 
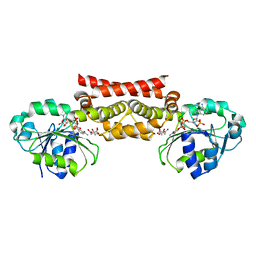 | | PREPHENATE DEHYDROGENASE FROM SOYBEAN | | 分子名称: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prephenate dehydrogenase 1 | | 著者 | Holland, C.K, Jez, J.M. | | 登録日 | 2017-07-18 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants.
Nat. Chem. Biol., 13, 2017
|
|
5WMH
 
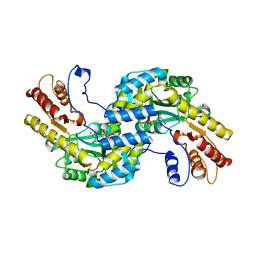 | | Arabidopsis thaliana prephenate aminotransferase | | 分子名称: | Bifunctional aspartate aminotransferase and glutamate/aspartate-prephenate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Holland, C.K, Jez, J.M. | | 登録日 | 2017-07-28 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for substrate recognition and inhibition of prephenate aminotransferase from Arabidopsis.
Plant J., 94, 2018
|
|
5WAX
 
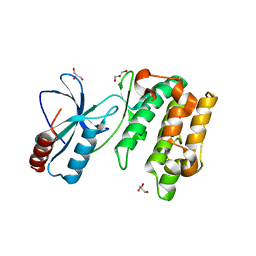 | | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10) | | 分子名称: | GLYCEROL, SAPK10 (serine/threonine-protein kinase 10) | | 著者 | Righetto, G.L, Counago, R.M, Halabelian, L, Santiago, A.S, Massirer, K.B, Arruda, P, Gileadi, O, Menossi, M, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | 登録日 | 2017-06-27 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10)
To Be Published
|
|
5ZNI
 
 | |
5WMK
 
 | |
