4CL0
 
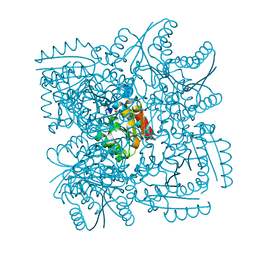 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | 分子名称: | (2R)-2-METHYL-3-DEHYDROQUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-10 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4CNO
 
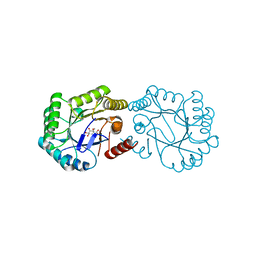 | | Structure of the Salmonella typhi Type I dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | 分子名称: | (2R)-2-METHYL-3-DEHYDROQUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-23 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Insights into substrate binding and catalysis in bacterial type I dehydroquinase.
Biochem. J., 462, 2014
|
|
4CKX
 
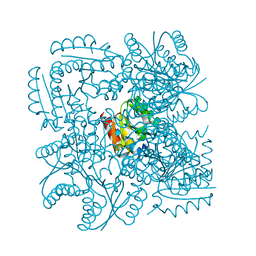 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase N12S mutant (Crystal Form 2) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-10 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4CNN
 
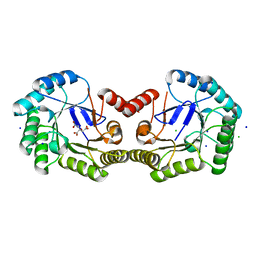 | | High resolution structure of Salmonella typhi type I dehydroquinase | | 分子名称: | 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-23 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Mechanistic Insight Into the Reaction Catalyzed by Type I Dehydroquinase
To be Published
|
|
1IW2
 
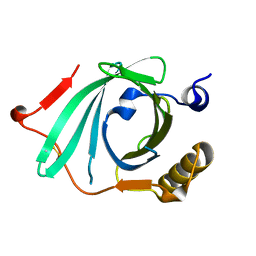 | | X-ray structure of Human Complement Protein C8gamma at pH=7.O | | 分子名称: | Complement Protein C8gamma | | 著者 | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | 登録日 | 2002-04-11 | | 公開日 | 2002-06-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
4CKY
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | 分子名称: | 2,2-dimethyl-3-dehydroquinic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-10 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4CLM
 
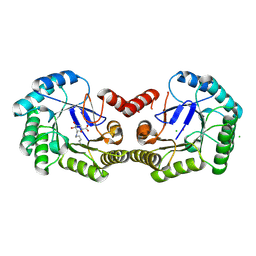 | | Structure of Salmonella typhi type I dehydroquinase irreversibly inhibited with a 1,3,4-trihydroxyciclohexane-1-carboxylic acid derivative | | 分子名称: | (1~{S},3~{S},4~{R},5~{R})-3-methyl-1,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Tizon, L, Maneiro, M, Lence, E, Poza, S, Lamb, H, Hawkins, A.R, Blanco, B, Sedes, A, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-15 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Irreversible covalent modification of type I dehydroquinase with a stable Schiff base.
Org. Biomol. Chem., 13, 2015
|
|
1IH2
 
 | |
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | 分子名称: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | 著者 | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | 登録日 | 2012-08-20 | | 公開日 | 2012-09-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
4CNP
 
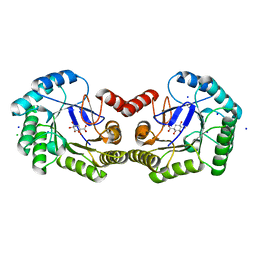 | | Structure of the Salmonella typhi type I dehydroquinase inhibited by a 3-epiquinic acid derivative | | 分子名称: | (2S)-2-HYDROXY-3-EPIQUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, CHLORIDE ION, ... | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-23 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Mechanistic Insight Into the Reaction Catalyzed by Type I Dehydroquinase
To be Published
|
|
1IH3
 
 | |
4CXA
 
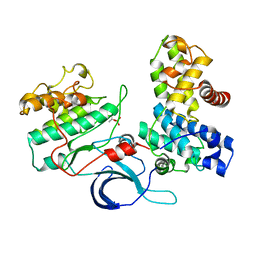 | | Crystal structure of the human CDK12-cyclin K complex bound to AMPPNP | | 分子名称: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Nowak, R, Goubin, S, Mahajan, R.P, Kopec, J, Froese, S, Tallant, C, Carpenter, E.P, Mackenzie, A, Faust, B, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2014-04-04 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
1IE9
 
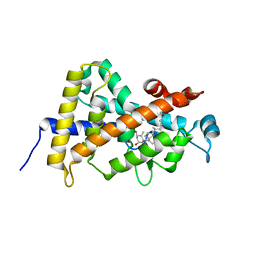 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | 分子名称: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | 著者 | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | 登録日 | 2001-04-09 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4CKZ
 
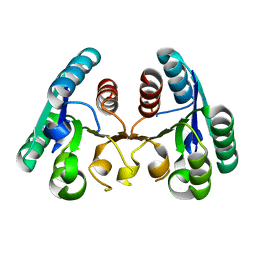 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant | | 分子名称: | 3-DEHYDROQUINATE DEHYDRATASE | | 著者 | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | 登録日 | 2014-01-10 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4CSA
 
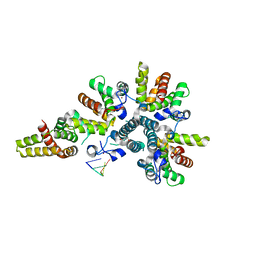 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to a DNA 4-mer | | 分子名称: | 5'-D(*AP*GP*TP*TP*AP)-3', GLYCEROL, M2-1, ... | | 著者 | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
1IQD
 
 | | Human Factor VIII C2 Domain complexed to human monoclonal BO2C11 Fab. | | 分子名称: | HUMAN FACTOR VIII, HUMAN MONOCLONAL BO2C11 FAB HEAVY CHAIN, HUMAN MONOCLONAL BO2C11 FAB LIGHT CHAIN | | 著者 | Spiegel Jr, P.C, Jacquemin, M, Saint-Remy, J.M, Stoddard, B.L, Pratt, K.P. | | 登録日 | 2001-07-21 | | 公開日 | 2001-08-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a factor VIII C2 domain-immunoglobulin G4kappa Fab complex: identification of an inhibitory antibody epitope on the surface of factor VIII.
Blood, 98, 2001
|
|
5QDC
 
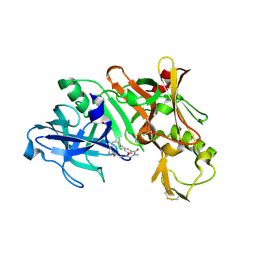 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | 分子名称: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1IDE
 
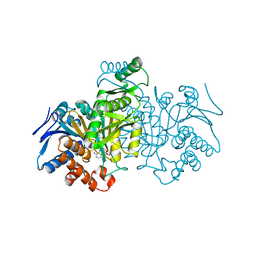 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | 分子名称: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | 著者 | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | 登録日 | 1995-01-18 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
4CVW
 
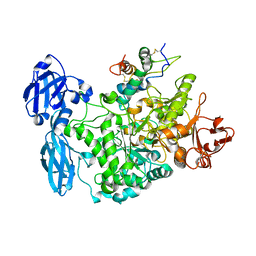 | | Structure of the barley limit dextrinase-limit dextrinase inhibitor complex | | 分子名称: | CALCIUM ION, LIMIT DEXTRINASE, LIMIT DEXTRINASE INHIBITOR | | 著者 | Moeller, M.S, Vester-Christensen, M.B, Jensen, J.M, Abou Hachem, M, Henriksen, A, Svensson, B. | | 登録日 | 2014-03-31 | | 公開日 | 2015-04-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal Structure of Barley Limit Dextrinase:Limit Dextrinase Inhibitor (Ld:Ldi) Complex Reveals Insights Into Mechanism and Diversity of Cereal-Type Inhibitors.
J.Biol.Chem., 290, 2015
|
|
5QD2
 
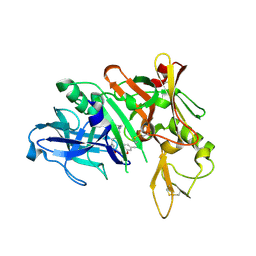 | | Crystal structure of BACE complex with BMC017 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1IH6
 
 | |
5QDA
 
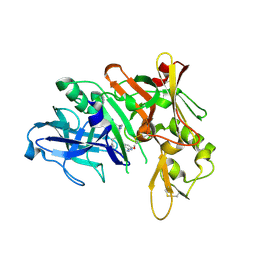 | | Crystal structure of BACE complex with BMC013 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
2N7B
 
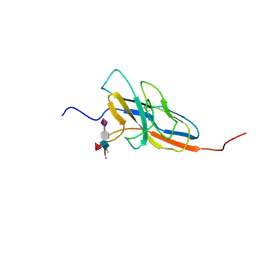 | | Solution structure of the human Siglec-8 lectin domain in complex with 6'sulfo sialyl Lewisx | | 分子名称: | 3-aminopropan-1-ol, N-acetyl-alpha-neuraminic acid-(2-3)-6-O-sulfo-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialic acid-binding Ig-like lectin 8 | | 著者 | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | 登録日 | 2015-09-07 | | 公開日 | 2016-07-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5QCO
 
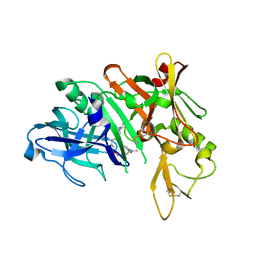 | | Crystal structure of BACE complex with BMC016 | | 分子名称: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1IDD
 
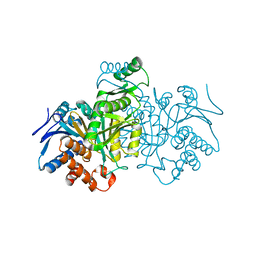 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT APO ENZYME | | 分子名称: | ISOCITRATE DEHYDROGENASE | | 著者 | Lee, M.E, Dyer, D.H, Klein, O.D, Bolduc, J.M, Stoddard, B.L, Koshland Junior, D.E. | | 登録日 | 1995-01-18 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
