6BHV
 
 | | Human PARP-1 bound to NAD+ analog benzamide adenine dinucleotide (BAD) | | Descriptor: | Poly [ADP-ribose] polymerase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylphenyl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Pascal, J.M, Langelier, M.F. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NAD+analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains.
Nat Commun, 9, 2018
|
|
2OV5
 
 | | Crystal structure of the KPC-2 carbapenemase | | Descriptor: | BICINE, Carbapenemase | | Authors: | Ke, W, Bethel, C.R, Thomson, J.M, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of KPC-2: Insights into Carbapenemase Activity in Class A beta-Lactamases.
Biochemistry, 46, 2007
|
|
5U9Q
 
 | | Ocellatin-LB1 | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, Santos, D.M, de Lima, M.E, Pilo-Veloso, D, Resende, J.M. | | Deposit date: | 2016-12-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
6BHT
 
 | | HIV-1 CA hexamer in complex with IP6, orthorhombic crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BQP
 
 | | Crystal Structure of the Human CAMKK2B in complex with Crenolanib | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, de Souza, G.P, dos Reis, C.V, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with Crenolanib
To Be Published
|
|
2P66
 
 | | Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*(3DR)P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Prasad, R, Batra, V.K, Yang, X.-P, Krahn, J.M, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the DNA Polymerase beta deoxyribose phosphate lyase mechanism
DNA REPAIR, 4, 2005
|
|
2P76
 
 | | Crystal structure of a Glycerophosphodiester Phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a Glycerophosphodiester Phosphodiesterase from Staphylococcus aureus
To be Published
|
|
6BHS
 
 | | HIV-1 CA hexamer in complex with IP6, hexagonal crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
2P8E
 
 | | Crystal structure of the serine/threonine phosphatase domain of human PPM1B | | Descriptor: | MAGNESIUM ION, PPM1B beta isoform variant 6 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Lau, C, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P61
 
 | | Crystal structure of protein TM1646 from Thermotoga maritima, Pfam DUF327 | | Descriptor: | Hypothetical protein TM_1646 | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein TM_1646 from Thermotoga maritima
To be Published
|
|
6BLE
 
 | | Crystal Structure of the Human CAMKK2B in complex with CP673451 | | Descriptor: | 1-{2-[5-(2-methoxyethoxy)-1H-benzimidazol-1-yl]quinolin-8-yl}piperidin-4-amine, Calcium/calmodulin-dependent protein kinase kinase 2, GLYCEROL | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with CP673451
To Be Published
|
|
2PAG
 
 | | Crystal structure of protein PSPTO_5518 from Pseudomonas syringae pv. tomato | | Descriptor: | CALCIUM ION, Hypothetical protein | | Authors: | Fedorov, A.A, Ramagopal, U, Toro, R, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Pseudomonas syringae pv. tomato.
To be Published
|
|
5UA7
 
 | | Ocellatin-LB2, solution structure in SDS micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB2 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
2OV2
 
 | | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J.M, Burgess-Brown, N, Bunkoczi, G, Debreczeni, J.E.D, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the human RAC3 in complex with the CRIB domain of human p21-activated kinase 4 (PAK4)
To be Published
|
|
6BYV
 
 | | Solution NMR structure of cysteine-rich calcium bound domains of very low density lipoprotein receptor | | Descriptor: | CALCIUM ION, Very low-density lipoprotein receptor | | Authors: | Banerjee, K, Gruschus, J.M, Tjandra, N, Yakovlev, S, Medved, L. | | Deposit date: | 2017-12-21 | | Release date: | 2018-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Recombinant Fragment Containing Three Fibrin-Binding Cysteine-Rich Domains of the Very Low Density Lipoprotein Receptor.
Biochemistry, 57, 2018
|
|
2P1J
 
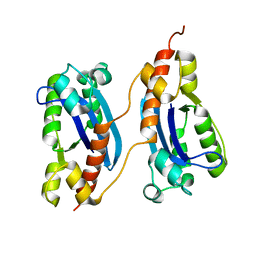 | | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima | | Descriptor: | DNA polymerase III polC-type | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Izuka, M, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a polC-type DNA polymerase III exonuclease domain from Thermotoga maritima
To be Published
|
|
5U9S
 
 | | Ocellatin-F1, solution structure in TFE by NMR spectroscopy | | Descriptor: | Ocellatin-F1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-F1
To Be Published
|
|
6BNZ
 
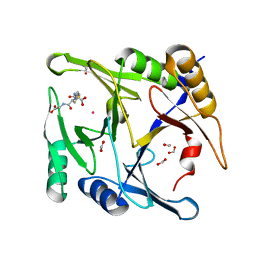 | | Crystal structure of E144Q-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
5U9Y
 
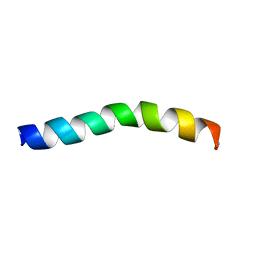 | | Ocellatin-F1 | | Descriptor: | Ocellatin-K1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
2P4M
 
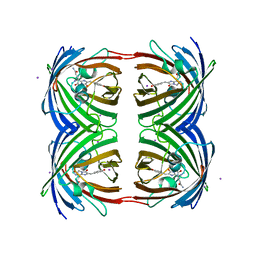 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
6BKU
 
 | | Crystal Structure of the Human CAMKK2B bound to GSK650394 | | Descriptor: | 2-cyclopentyl-4-(5-phenyl-1H-pyrrolo[2,3-b]pyridin-3-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2, FORMIC ACID | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B bound to GSK650394
To Be Published
|
|
6BPZ
 
 | | Structure of the mechanically activated ion channel Piezo1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1,mouse Piezo1,Piezo-type mechanosensitive ion channel component 1,Piezo-type mechanosensitive ion channel component 1 | | Authors: | Saotome, K, Kefauver, J.M, Patapoutian, A, Ward, A.B. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the mechanically activated ion channel Piezo1.
Nature, 554, 2018
|
|
6BRC
 
 | | Crystal Structure of the Human CAMKK2B in complex with AP26113-analog (ALK-IN-1) | | Descriptor: | 5-chloro-N~2~-{4-[4-(dimethylamino)piperidin-1-yl]-2-methoxyphenyl}-N~4~-[2-(dimethylphosphoryl)phenyl]pyrimidine-2,4-diamine, Calcium/calmodulin-dependent protein kinase kinase 2, SODIUM ION | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-30 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with AP26113-analog (ALK-IN-1)
To Be Published
|
|
6BRU
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a (S)-2-phenylaminopteridinone inhibitor | | Descriptor: | (7S)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7,8-trimethyl-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Aquino, B, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a (S)-2-phenylaminopteridinone inhibitor
To Be Published
|
|
6BT5
 
 | | Human mGlu8 Receptor complexed with L-AP4 | | Descriptor: | (2S)-2-amino-4-phosphonobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 8 | | Authors: | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | Deposit date: | 2017-12-05 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
