1M7T
 
 | |
1RRI
 
 | | DHNA complex with 3-(5-amino-7-hydroxy-[1,2,3] triazolo [4,5-d]pyrimidin-2-yl)-benzoic acid | | Descriptor: | 3-(5-AMINO-7-HYDROXY-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-2-YL)-BENZOIC ACID, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1M1J
 
 | | Crystal structure of native chicken fibrinogen with two different bound ligands | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, Z, Kollman, J.M, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Native Chicken Fibrinogen at 2.7 A Resolution
Biochemistry, 40, 2001
|
|
4BLT
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S292A MUTANT IN COMPLEX WITH AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
3EBO
 
 | | Glycogen Phosphorylase b/Chrysin complex | | Descriptor: | Glycogen phosphorylase, muscle form, chrysin | | Authors: | Oikonomakos, N.G, Zographos, S.E, Leonidas, D.D, Hayes, J.M, Tiraidis, C, Alexacou, K.-M. | | Deposit date: | 2008-08-28 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sourcing the affinity of flavonoids for the glycogen phosphorylase inhibitor site via crystallography, kinetics and QM/MM-PBSA binding studies: Comparison of chrysin and flavopiridol
Food Chem.Toxicol., 61, 2013
|
|
3P6G
 
 | |
3EOI
 
 | | CRYSTAL STRUCTURE OF putative PROTEIN PilM from Escherichia coli B7A | | Descriptor: | PilM | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J.B, Sauder, J.M, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
1ROC
 
 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
2HIK
 
 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
1R48
 
 | | Solution structure of the C-terminal cytoplasmic domain residues 468-497 of Escherichia coli protein ProP | | Descriptor: | Proline/betaine transporter | | Authors: | Zoetewey, D.L, Tripet, B.P, Kutateladze, T.G, Overduin, M.J, Wood, J.M, Hodges, R.S. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Antiparallel Coiled-coil Domain from Escherichia coli Osmosensor ProP.
J.Mol.Biol., 334, 2003
|
|
3GH8
 
 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) bound to FMN and di-iodotyrosine (DIT) | | Descriptor: | 3,5-DIIODOTYROSINE, FLAVIN MONONUCLEOTIDE, Iodotyrosine dehalogenase 1, ... | | Authors: | Thomas, S.R, McTamney, P.M, Adler, J.M, LaRonde-LeBlanc, N, Rokita, S.E. | | Deposit date: | 2009-03-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
J.Biol.Chem., 284, 2009
|
|
1ROS
 
 | | Crystal structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-[2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL)ETHYL]-4-(4'-ETHOXY-1,1'-BIPHENYL-4-YL)-4-OXOBUTANOIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J.Mol.Biol., 341, 2004
|
|
3GHY
 
 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
3EEY
 
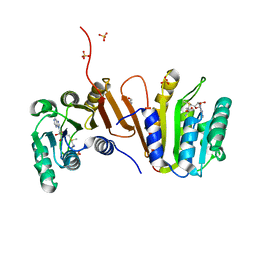 | | CRYSTAL STRUCTURE OF PUTATIVE RRNA-METHYLASE FROM Clostridium thermocellum | | Descriptor: | GLYCEROL, Putative rRNA methylase, S-ADENOSYLMETHIONINE, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Bain, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rrna-Methylase from Clostridium Thermocellum
To be Published
|
|
4C9W
 
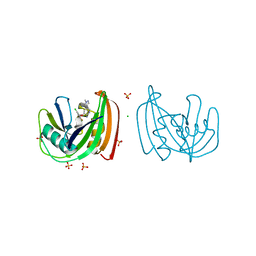 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
1R7I
 
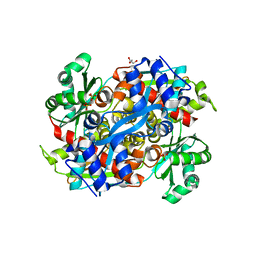 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
3E61
 
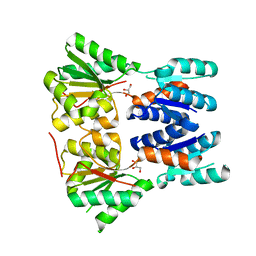 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
3OLM
 
 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
4BIU
 
 | | Crystal structure of CpxAHDC (orthorhombic form 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SENSOR PROTEIN CPXA, SULFATE ION | | Authors: | Mechaly, A.E, Sassoon, N, Betton, J.M, Alzari, P.M. | | Deposit date: | 2013-04-13 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Segmental Helical Motions and Dynamical Asymmetry Modulate Histidine Kinase Autophosphorylation.
Plos Biol., 12, 2014
|
|
4BD7
 
 | | Bax domain swapped dimer induced by octylmaltoside | | Descriptor: | APOPTOSIS REGULATOR BAX, CHLORIDE ION, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
1SM2
 
 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
4BIZ
 
 | |
2FZ6
 
 | | Crystal structure of hydrophobin HFBI | | Descriptor: | Hydrophobin-1, ZINC ION | | Authors: | Hakanpaa, J.M, Rouvinen, J. | | Deposit date: | 2006-02-09 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of Trichoderma reesei hydrophobin HFBI--The structure of a protein amphiphile with and without detergent interaction.
Protein Sci., 15, 2006
|
|
3P8H
 
 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
4BD2
 
 | | Bax domain swapped dimer in complex with BidBH3 | | Descriptor: | APOPTOSIS REGULATOR BAX, BH3-INTERACTING DOMAIN DEATH AGONIST | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-04 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
