2HHQ
 
 | | O6-methyl-guanine:T pair in the polymerase-10 basepair position | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*AP*TP*GP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6IF1
 
 | | Crystal structure of Ube2K and K48-linked di-ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Lee, J.-G, Youn, H.-S, Lee, Y, An, J.Y, Park, K.R, Kang, J.Y, Lim, J.J, Eom, S.H. | | Deposit date: | 2018-09-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.466 Å) | | Cite: | Crystal structure of the Ube2K/E2-25K and K48-linked di-ubiquitin complex provides structural insight into the mechanism of K48-specific ubiquitin chain synthesis.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
2HHX
 
 | | O6-methyl-guanine in the polymerase template preinsertion site | | Descriptor: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2LXL
 
 | | Lip5(mit)2 | | Descriptor: | Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
2HVH
 
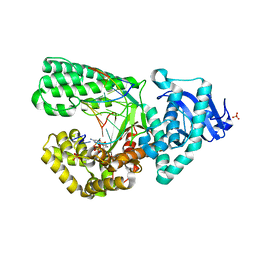 | | ddCTP:O6MeG pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*CP*A*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HVI
 
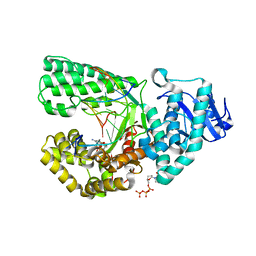 | | ddCTP:G pair in the polymerase active site (0 position) | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*C*AP*TP*GP*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*(DDG))-3', ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2MIT
 
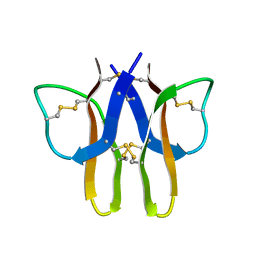 | |
2HW3
 
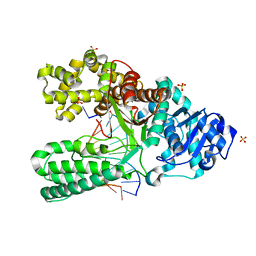 | | T:O6-methyl-guanine pair in the polymerase postinsertion site (-1 basepair position) | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*T)-3', 5'-D(*GP*TP*A*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA Polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-07-31 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HTW
 
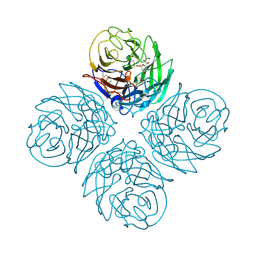 | | N4 neuraminidase in complex with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
6JPU
 
 | | CryoEM structure of Abo1 hexamer - apo complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6JRP
 
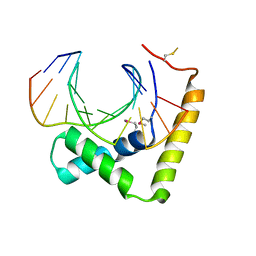 | | Crystal structure of CIC-HMG-ETV5-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*AP*TP*GP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*CP*AP*TP*TP*CP*AP*T)-3'), Protein capicua homolog | | Authors: | Song, J.J, Lee, H. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of Capicua HMG-box domain complexed with the ETV5-DNA and its implications for Capicua-mediated cancers.
Febs J., 286, 2019
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
2JJY
 
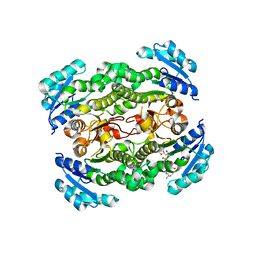 | | Crystal structure of Francisella tularensis enoyl reductase (ftFabI) with bound NAD | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Luckner, S.R, Lu, H, Truglio, J.J, Tonge, P.J, Kisker, C. | | Deposit date: | 2008-04-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Slow-Onset Inhibition of the Fabi Enoyl Reductase from Francisella Tularensis: Residence Time and in Vivo Activity
Acs Chem.Biol., 4, 2009
|
|
2JH2
 
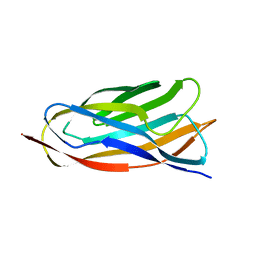 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2JZ3
 
 | | SOCS box elonginBC ternary complex | | Descriptor: | Suppressor of cytokine signaling 3, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | Authors: | Babon, J.J, Sabo, J, Soetopo, A, Yao, S, Bailey, M.F, Zhang, J, Nicola, N.A, Norton, R.S. | | Deposit date: | 2007-12-27 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase
J.Mol.Biol., 381, 2008
|
|
2JQK
 
 | | VPS4B MIT-CHMP2B Complex | | Descriptor: | Charged multivesicular body protein 2b, Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JWW
 
 | |
2K4F
 
 | | Mouse CD3epsilon Cytoplasmic Tail | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Xu, C, Call, M.E, Schwieters, C.D, Schnell, J.R, Gagnon, E.E, Wucherpfennig, K.W, Chou, J.J. | | Deposit date: | 2008-06-07 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Regulation of T Cell Receptor Activation by Dynamic Membrane Binding of the CD3varepsilon Cytoplasmic Tyrosine-Based Motif.
Cell(Cambridge,Mass.), 135, 2008
|
|
6KVO
 
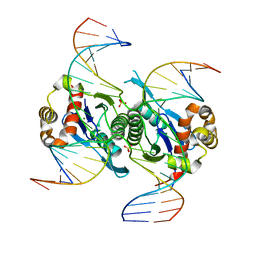 | | Crystal structure of chloroplast resolvase in complex with Holliday junction | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*TP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*TP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*TP*TP*GP*T)-3'), ... | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-09-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6LCM
 
 | | Crystal structure of chloroplast resolvase ZmMOC1 with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ZmMoc1 | | Authors: | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2019-11-19 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
2OT5
 
 | |
2IBX
 
 | | Influenza virus (VN1194) H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yamada, S, Russell, R.J, Gamblin, S.J, Skehel, J.J, Kawaoka, Y. | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors.
Nature, 444, 2006
|
|
2P4L
 
 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | Descriptor: | I-superfamily conotoxin r11a | | Authors: | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2007-03-12 | | Release date: | 2007-09-25 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
2HU0
 
 | | N1 neuraminidase in complex with oseltamivir 1 | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
