1FWX
 
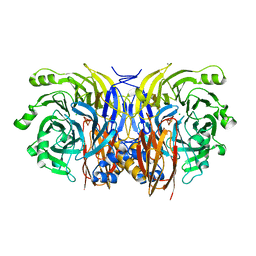 | | CRYSTAL STRUCTURE OF NITROUS OXIDE REDUCTASE FROM P. DENITRIFICANS | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Brown, K, Djinovic-Carugo, K, Haltia, T, Cabrito, I, Saraste, M, Moura, J.J, Moura, I, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-09-25 | | Release date: | 2001-09-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Revisiting the Catalytic CuZ Cluster of Nitrous Oxide (N2O) Reductase. Evidence of a Bridging Inorganic Sulfur
J.Biol.Chem., 275, 2000
|
|
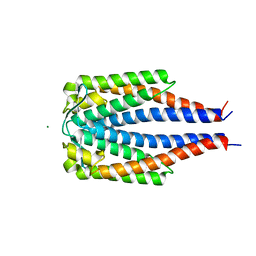
1GGQ
 
 | |
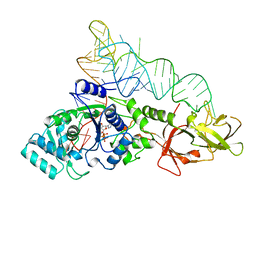
1GTR
 
 | |
1G88
 
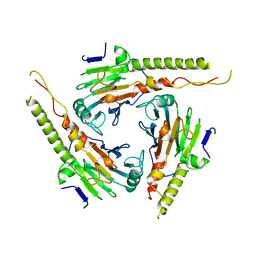 | | S4AFL3ARG515 MUTANT | | Descriptor: | SMAD4 | | Authors: | Chako, B.M, Qin, B, Lam, S.S, Correia, J.J, Lin, K. | | Deposit date: | 2000-11-16 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization.
Nat.Struct.Biol., 8, 2001
|
|
1HA0
 
 | | HEMAGGLUTININ PRECURSOR HA0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HEMAGGLUTININ PRECURSOR), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, J, Ho Lee, K, Steinhauer, D.A, Stevens, D.J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1998-10-08 | | Release date: | 1998-10-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation.
Cell(Cambridge,Mass.), 95, 1998
|
|
1F1M
 
 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | Descriptor: | OUTER SURFACE PROTEIN C, ZINC ION | | Authors: | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | Deposit date: | 2000-05-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
1FR1
 
 | |
1HCJ
 
 | |
1GVZ
 
 | | Prostate Specific Antigen (PSA) from stallion seminal plasma | | Descriptor: | ACETATE ION, GLYCEROL, KALLIKREIN-1E2 | | Authors: | Carvalho, A.L, Sanz, L, Barettino, D, Romero, A, Calvete, J.J, Romao, M.J. | | Deposit date: | 2002-02-28 | | Release date: | 2002-09-12 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of a Prostate Kallikrein Isolated from Stallion Seminal Plasma: A Homologue of Human Psa
J.Mol.Biol., 322, 2002
|
|
2LL0
 
 | | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans | | Descriptor: | Uncharacterized protein | | Authors: | Serrano, P, Geralt, M, Wuthrich, K, Morales-Rios, E, Zarco-Zavala, M, Garcia-Trejo, J.J, Dutta, S.K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the putative ATPase regulatory protein YP_916642.1 from Paracoccus denitrificans
To be Published
|
|
5XBK
 
 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4, histone H3 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.223 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
5KEK
 
 | | Structure Determination of a Self-Assembling DNA Crystal | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, Birktoft, J.J, Seeman, N.C, Yan, H. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Construction and Structure Determination of a Three-Dimensional DNA Crystal.
J.Am.Chem.Soc., 138, 2016
|
|
5KF6
 
 | |
6OL0
 
 | | Structure of VcINDY bound to Malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SODIUM ION, Transporter, ... | | Authors: | Sauer, D.B, Marden, J.J, Wang, D.N. | | Deposit date: | 2019-04-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Elevator mechanism dynamics in a sodium-coupled dicarboxylate transporter
Biorxiv, 2022
|
|
5K8S
 
 | | cAMP bound PfPKA-R (297-441) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CAMP-dependent protein kinase regulatory subunit | | Authors: | Littler, D.R, Gilson, P.R, Crabb, B.S, Rossjohn, J.J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Disrupting the Allosteric Interaction between the Plasmodium falciparum cAMP-dependent Kinase and Its Regulatory Subunit.
J. Biol. Chem., 291, 2016
|
|
5KEO
 
 | | Structure Determination of a Self-Assembling DNA Crystal | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, Birktoft, J.J, Seeman, N.C, Yan, H. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Construction and Structure Determination of a Three-Dimensional DNA Crystal.
J.Am.Chem.Soc., 138, 2016
|
|
6O5G
 
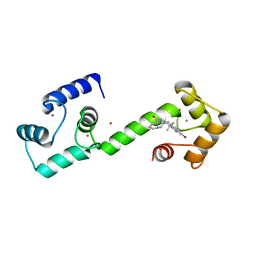 | | Calmodulin in complex with isomalbrancheamide D | | Descriptor: | (5aS,12aS,13aS)-9-bromo-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7 ,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2019-03-02 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2LQA
 
 | |
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6O4C
 
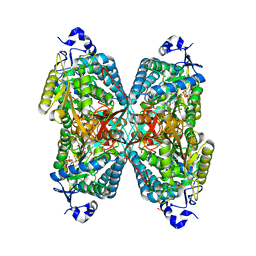 | | Structure of ALDH7A1 mutant W175A complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4K
 
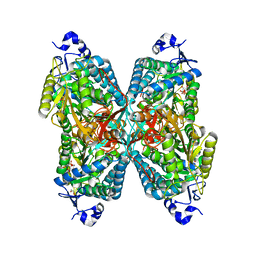 | | Structure of ALDH7A1 mutant E399Q complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
5KBZ
 
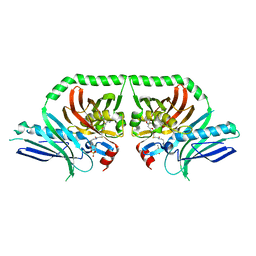 | | Structure of the PksA Product Template domain in complex with a phosphopantetheine mimetic | | Descriptor: | (14R)-14-hydroxy-15,15-dimethyl-1-[5-({[(5-methyl-1,2-oxazol-3-yl)methyl]sulfanyl}methyl)-1,2-oxazol-3-yl]-4,9,13-trioxo-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen phosphate, Noranthrone synthase | | Authors: | Tsai, S.C, Burkart, M.D, Townsend, C.A, Barajas, J.F, Shakya, G, Topper, C.L, Moreno, G, Jackson, D.R, Rivera, H, La Clair, J.J, Vagstad, A. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Polyketide mimetics yield structural and mechanistic insights into product template domain function in nonreducing polyketide synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2LVJ
 
 | | solution structure of hemi-Mg-bound Phl p 7 | | Descriptor: | MAGNESIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
6O4I
 
 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6OL1
 
 | |
