1M76
 
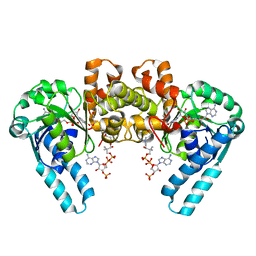 | |
1MD9
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB AND AMP | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MPX
 
 | | ALPHA-AMINO ACID ESTER HYDROLASE LABELED WITH SELENOMETHIONINE | | Descriptor: | CALCIUM ION, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Hensgens, C.M.H, de Vries, E.J, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2002-09-13 | | Release date: | 2003-04-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The sequence and crystal structure of the alpha-amino acid ester hydrolase from Xanthomonas citri define a new family of beta-lactam antibiotic acylases.
J.Biol.Chem., 278, 2003
|
|
1MLD
 
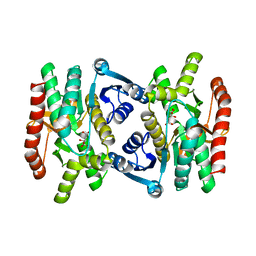 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
2RBF
 
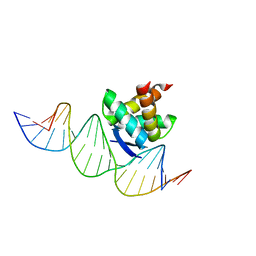 | | Structure of the ribbon-helix-helix domain of Escherichia coli PutA (PutA52) complexed with operator DNA (O2) | | Descriptor: | Bifunctional protein putA, DNA (5'-D(*DTP*DT*DTP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DCP*DAP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DGP*DAP*DAP*DAP*DGP*DGP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DAP*DAP*DA)-3') | | Authors: | Tanner, J.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA.
J.Mol.Biol., 381, 2008
|
|
8PD0
 
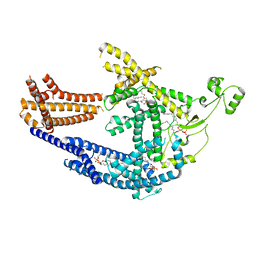 | | cryo-EM structure of Doa10 in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PDA
 
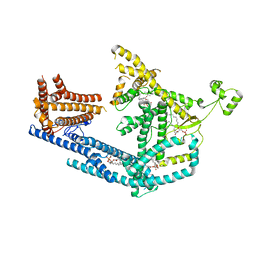 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
1M6O
 
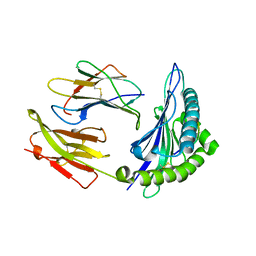 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1MIT
 
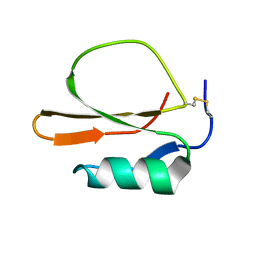 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
7KAA
 
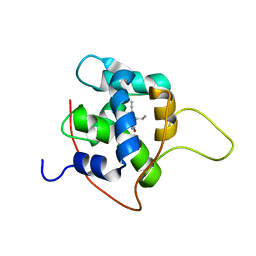 | | NMR solution structures of tirasemtiv drug bound to a fast skeletal troponin C-troponin I complex | | Descriptor: | 6-ethynyl-1-(pentan-3-yl)-1H-imidazo[4,5-b]pyrazin-2-ol, CALCIUM ION, Troponin C, ... | | Authors: | Mercier, P, Li, M.X, Hartman, J.J, Sykes, B.D. | | Deposit date: | 2020-09-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Tirasemtiv Activation of Fast Skeletal Muscle.
J.Med.Chem., 64, 2021
|
|
1M6P
 
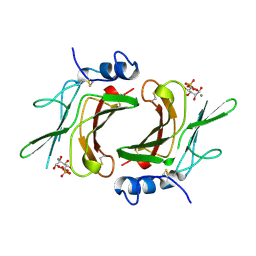 | | EXTRACYTOPLASMIC DOMAIN OF BOVINE CATION-DEPENDENT MANNOSE 6-PHOSPHATE RECEPTOR | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, CATION-DEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, MANGANESE (II) ION | | Authors: | Roberts, D.L, Weix, D.J, Dahms, N.M, Kim, J.J.-P. | | Deposit date: | 1998-04-19 | | Release date: | 1999-04-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of lysosomal enzyme recognition: three-dimensional structure of the cation-dependent mannose 6-phosphate receptor.
Cell(Cambridge,Mass.), 93, 1998
|
|
7JVE
 
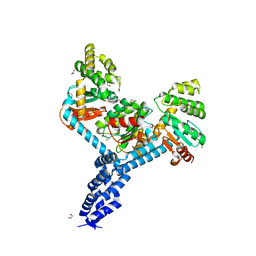 | | Crystal structure of Salmonella enterica Typhimurium BcfH | | Descriptor: | 1,2-ETHANEDIOL, DsbA family protein, MAGNESIUM ION, ... | | Authors: | Subedi, P, Heras, B, Hor, L, Paxman, J.J. | | Deposit date: | 2020-08-21 | | Release date: | 2021-04-21 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Salmonella enterica BcfH Is a Trimeric Thioredoxin-Like Bifunctional Enzyme with Both Thiol Oxidase and Disulfide Isomerase Activities.
Antioxid.Redox Signal., 35, 2021
|
|
1MJS
 
 | |
1MHI
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF AN INSULIN DIMER. A STUDY OF THE B9(ASP) MUTANT OF HUMAN INSULIN USING NUCLEAR MAGNETIC RESONANCE DISTANCE GEOMETRY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | INSULIN | | Authors: | Jorgensen, A.M.M, Kristensen, S.M, Led, J.J, Balschmidt, P. | | Deposit date: | 1994-11-30 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of an insulin dimer. A study of the B9(Asp) mutant of human insulin using nuclear magnetic resonance, distance geometry and restrained molecular dynamics.
J.Mol.Biol., 227, 1992
|
|
8OQ2
 
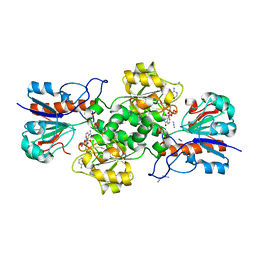 | | Binding of NADP to a formate dehydrogenase from Starkeya novella. | | Descriptor: | AZIDE ION, Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Partipilo, M, Whittaker, J.J, Pontillo, N, Guskov, A, Slotboom, D.J. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of NADP to a formate dehydrogenase from Starkeya novella.
To Be Published
|
|
2R80
 
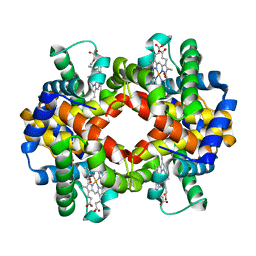 | | Pigeon Hemoglobin (OXY form) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Ponnuswamy, M.N, Packianathan, C, Sundaresan, S, Neelagandan, K, Palani, K, Muller, J.J, Heinemann, U. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-ray crystal structure analysis of Hemolgobin from Pigeon (Columba Livia) at 1.44 angstrom
To be Published
|
|
1M1Q
 
 | | P222 oxidized structure of the tetraheme cytochrome c from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
2RLF
 
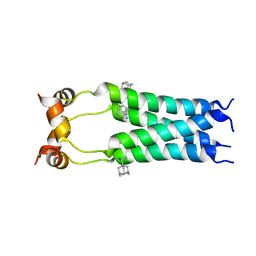 | |
2R83
 
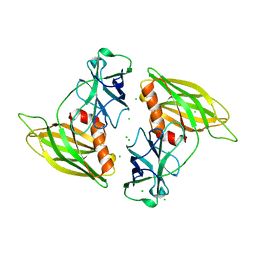 | | Crystal structure analysis of human synaptotagmin 1 C2A-C2B | | Descriptor: | CHLORIDE ION, Synaptotagmin-1 | | Authors: | Sutton, R.B, Fuson, K.L, Montes, M, Robert, J.J. | | Deposit date: | 2007-09-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association.
Biochemistry, 46, 2007
|
|
2TPS
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | MAGNESIUM ION, PROTEIN (THIAMIN PHOSPHATE SYNTHASE), PYROPHOSPHATE 2-, ... | | Authors: | Chiu, H.-J, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution.
Biochemistry, 38, 1999
|
|
8P3C
 
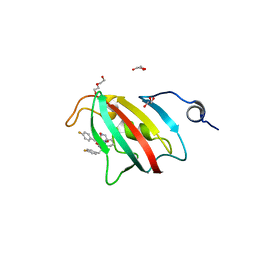 | | Full length structure of BpMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P3D
 
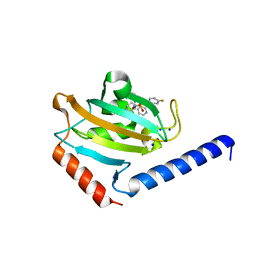 | | Full length structure of TcMIP with bound inhibitor NJS224. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-(pyridin-3-ylmethylamino)pentan-2-yl]piperidine-2-carboxamide, SODIUM ION, peptidylprolyl isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
8P42
 
 | | Full length structure of TcMIP with bound inhibitor NJS227. | | Descriptor: | (2~{S})-1-[(4-fluorophenyl)methylsulfonyl]-~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-(pyridin-3-ylmethylamino)propan-2-yl]piperidine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Macrophage infectivity potentiator | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2023-05-19 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural dynamics of macrophage infectivity potentiator proteins (MIPs) are differentially modulated by inhibitors and appendage domains
To Be Published
|
|
7L0R
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
