3J41
 
 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4AD4
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine and alpha-1,2- mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A54
 
 | | Structural basis of the Dcp1:Dcp2 mRNA decapping complex activation by Edc3 and Scd6 | | Descriptor: | EDC3, MRNA DECAPPING COMPLEX SUBUNIT 2 | | Authors: | Fromm, S.A, Truffault, V, Kamenz, J, Braun, J.E, Hoffmann, N.A, Izaurralde, E, Sprangers, R. | | Deposit date: | 2011-10-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Edc3- and Scd6-Mediated Activation of the Dcp1:Dcp2 Mrna Decapping Complex.
Embo J., 31, 2011
|
|
429D
 
 | |
2ODB
 
 | | The crystal structure of human cdc42 in complex with the CRIB domain of human p21-activated kinase 6 (PAK6) | | Descriptor: | CHLORIDE ION, Human Cell Division Cycle 42 (CDC42), MAGNESIUM ION, ... | | Authors: | Ugochukwu, E, Yang, X, Elkins, J, Soundararajan, M, Pike, A.C.W, Eswaran, J, Burgess, N, Debreczeni, J.E, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Gileadi, O, von Delft, F, Knapp, S, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of human cdc42 in complex with the CRIB domain of human p21-activated kinase 6 (PAK6)
To be Published
|
|
4AD2
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4CZZ
 
 | | Histone demethylase LSD1(KDM1A)-CoREST3 Complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 3 | | Authors: | Barrios, A.P, Gomez, A.V, Saez, J.E, Ciossani, G, Toffolo, E, Battaglioli, E, Mattevi, A, Andres, M.E. | | Deposit date: | 2014-04-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential Properties of Transcriptional Complexes Formed by the Corest Family.
Mol.Cell.Biol., 34, 2014
|
|
7U55
 
 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin at pH 4.5 | | Descriptor: | CHLORIDE ION, DODECANE, Heliorhodopsin, ... | | Authors: | Besaw, J.E, De Guzman, P, Miller, R.J.D, Ernst, O.P. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Low pH structure of heliorhodopsin reveals chloride binding site and intramolecular signaling pathway.
Sci Rep, 12, 2022
|
|
5NXB
 
 | | Mouse galactocerebrosidase in complex with saposin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Graham, S.C, Hill, C.H, Deane, J.E. | | Deposit date: | 2017-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The mechanism of glycosphingolipid degradation revealed by a GALC-SapA complex structure.
Nat Commun, 9, 2018
|
|
2OME
 
 | | Crystal structure of human CTBP2 dehydrogenase complexed with NAD(H) | | Descriptor: | C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pilka, E.S, Guo, K, Rojkova, A, Debreczeni, J.E, Kavanagh, K.L, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-22 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human CTBP2 dehydrogenase complexed with NAD(H)
To be Published
|
|
2YFU
 
 | | CBM62 FROM CLOSTRIDIUM THERMOCELLUM XYL5A | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6, GLYCEROL | | Authors: | Montanier, C.Y, Correia, M.A.S, Flint, J.E, Zhu, Y, Basle, A, Mckee, L.S, Prates, J.A.M, Polizzi, S.J, Coutinho, P.M, Henrissat, B, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2011-04-08 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel, Noncatalytic Carbohydrate-Binding Module Displays Specificity for Galactose-Containing Polysaccharides Through Calcium-Mediated Oligomerization.
J.Biol.Chem., 286, 2011
|
|
5NZO
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 1-methyl-3-phenyl-1H-pyrazol-5-amine | | Descriptor: | 2-methyl-5-phenyl-pyrazol-3-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5O89
 
 | | Crystal Structure of rsEGFP2 in the fluorescent on-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O8C
 
 | | Composite structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
3GS8
 
 | | An all-RNA hairpin ribozyme A38N1dA38 variant with a transition-state mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
2L27
 
 | | NMR Structure of the ECD1 of CRF-R1 in complex with a peptide agonist | | Descriptor: | Corticotropin-releasing factor receptor 1, peptide agonist | | Authors: | Grace, C.R.R, Perrin, M.H, Gulyas, J.R.R, Rivier, J.E, Vale, W.W, Riek, R.R. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-01 | | Last modified: | 2017-03-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the first extracellular domain of corticotropin-releasing factor receptor 1 (ECD1-CRF-R1) complexed with a high affinity agonist.
J.Biol.Chem., 285, 2010
|
|
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
2L5S
 
 | | Solution structure of the extracellular domain of the TGF-beta type I receptor | | Descriptor: | TGF-beta receptor type-1 | | Authors: | Zuniga, J.E, Ilangovan, U, Pardeep, M, Hinck, C, Huang, T. | | Deposit date: | 2010-11-04 | | Release date: | 2011-10-26 | | Method: | SOLUTION NMR | | Cite: | The TbetaR-I Pre-Helix Extension Is Structurally Ordered in the Unbound Form and Its Flanking Prolines Are Essential for Binding
J.Mol.Biol., 412, 2011
|
|
1LJ7
 
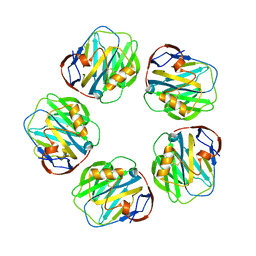 | | Crystal structure of calcium-depleted human C-reactive protein from perfectly twinned data | | Descriptor: | C-reactive protein | | Authors: | Ramadan, M.A, Shrive, A.K, Holden, D, Myles, D.A, Volanakis, J.E, DeLucas, L.J, Greenhough, T.J. | | Deposit date: | 2002-04-19 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The three-dimensional structure of calcium-depleted human C-reactive protein from perfectly twinned crystals.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2ZFC
 
 | | X-ray crystal structure of an engineered N-terminal HIV-1 GP41 trimer with enhanced stability and potency | | Descriptor: | HIV-1 GP41 | | Authors: | Dwyer, J.J, Wilson, K.L, Martin, K, Seedorff, J.E, Hasan, A, Kim, H. | | Deposit date: | 2007-12-29 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency
Protein Sci., 17, 2008
|
|
4CCD
 
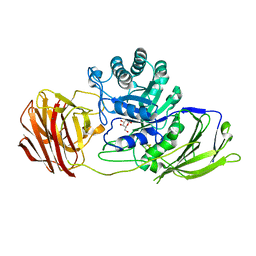 | | STRUCTURE OF MOUSE GALACTOCEREBROSIDASE WITH D-GALACTAL: ENZYME- INTERMEDIATE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Hill, C.H, Graham, S.C, Read, R.J, Deane, J.E. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Snapshots Illustrate the Catalytic Cycle of Beta-Galactocerebrosidase, the Defective Enzyme in Krabbe Disease
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2LKR
 
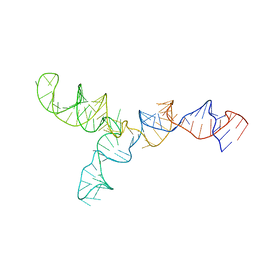 | | Yeast U2/U6 complex | | Descriptor: | RNA (111-MER) | | Authors: | Burke, J.E, Sashital, D.G, Zuo, X, Wang, Y, Butcher, S.E. | | Deposit date: | 2011-10-19 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast U2/U6 snRNA complex.
Rna, 18, 2012
|
|
5OD5
 
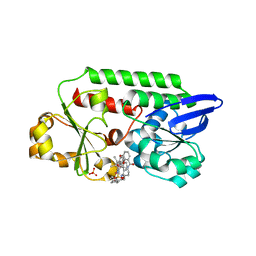 | | Periplasmic binding protein CeuE complexed with a synthetic catalyst | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 4-(aminomethyl)-~{N}-(pyridin-2-ylmethyl)benzenesulfonamide, Azotochelin, ... | | Authors: | Duhme-Klair, A.K, Raines, D.J, Clarke, J.E, Blagova, E.V, Dodson, E.J, Wilson, K.S. | | Deposit date: | 2017-07-04 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox-switchable siderophore anchor enables reversible artificial metalloenzyme assembly
Nat Catal, 2018
|
|
3GCA
 
 | |
1MQW
 
 | | Structure of the MT-ADPRase in complex with three Mn2+ ions and AMPCPR, a Nudix enzyme | | Descriptor: | ADPR pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MANGANESE (II) ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
