7R9Y
 
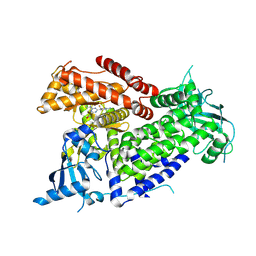 | | Structure of PIK3CA with covalent inhibitor 22 | | Descriptor: | N-[2-(4-{4-[2-amino-4-(difluoromethyl)pyrimidin-5-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}piperazin-1-yl)-2-oxoethyl]-N-methyl-1-(prop-2-enoyl)piperidine-4-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Burke, J.E, McPhail, J.A. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Covalent Proximity Scanning of a Distal Cysteine to Target PI3K alpha.
J.Am.Chem.Soc., 144, 2022
|
|
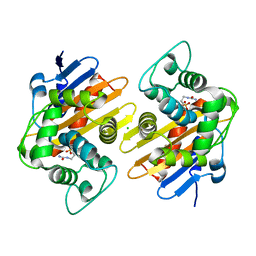
7LXG
 
 | |
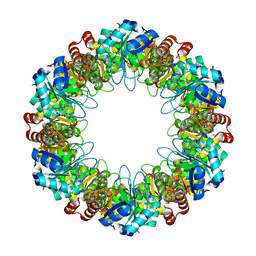
7LB5
 
 | |
7LB6
 
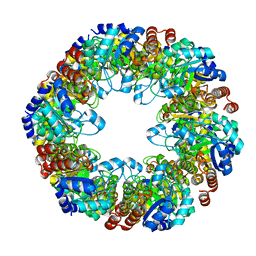 | | PDX1.2/PDX1.3 co-expression complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2 | | Authors: | Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Tunable Heteroassembly of a Plant Pseudoenzyme-Enzyme Complex.
Acs Chem.Biol., 16, 2021
|
|
5UQY
 
 | | Crystal structure of Marburg virus GP in complex with the human survivor antibody MR78 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP1, ... | | Authors: | Hashiguchi, T, Fusco, M.L, Hastie, K.M, Bomholdt, Z.A, Lee, J.E, Flyak, A.I, Matsuoka, R, Kohda, D, Yanagi, Y, Hammel, M, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for Marburg virus neutralization by a cross-reactive human antibody.
Cell, 160, 2015
|
|
7MJS
 
 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
7MEZ
 
 | | Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 5 | | Authors: | Burke, J.E, Dalwadi, U, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the phosphoinositide 3-kinase (PI3K) p110 gamma-p101 complex reveals molecular mechanism of GPCR activation.
Sci Adv, 7, 2021
|
|
2AAI
 
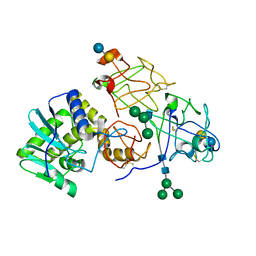 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
7SEP
 
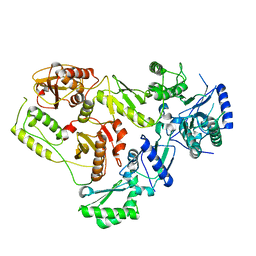 | |
1A54
 
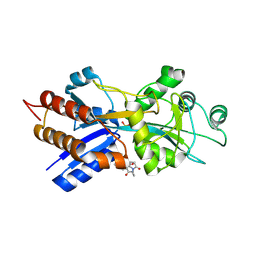 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C LABELLED WITH A COUMARIN FLUOROPHORE AND BOUND TO DIHYDROGENPHOSPHATE ION | | Descriptor: | DIHYDROGENPHOSPHATE ION, N-[2-(1-MALEIMIDYL)ETHYL]-7-DIETHYLAMINOCOUMARIN-3-CARBOXAMIDE, Phosphate-binding protein PstS | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
1A55
 
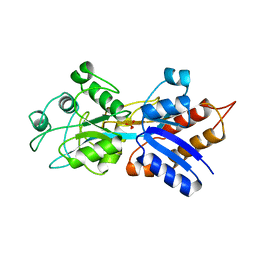 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C | | Descriptor: | DIHYDROGENPHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
7SJX
 
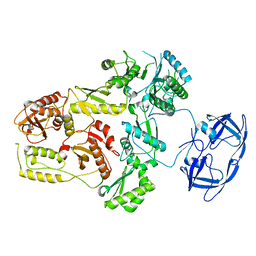 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
8A41
 
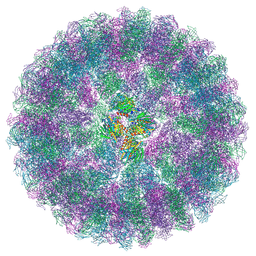 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A3C
 
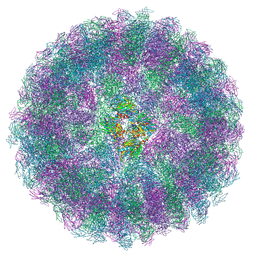 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
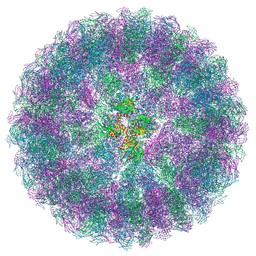 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
2BYL
 
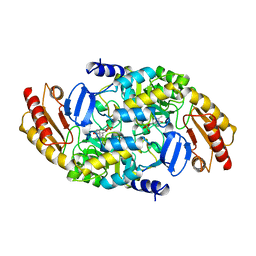 | | Structure of ornithine aminotransferase triple mutant Y85I Y55A G320F | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Markova, M, Peneff, C, Hewlins, M.J.E, Schirmer, T, John, R.A. | | Deposit date: | 2005-08-03 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Determinants of Substrate Specificity in Omega-Aminotransferases.
J.Biol.Chem., 280, 2005
|
|
7ZPC
 
 | | CDK2 in complex 9K-DOS | | Descriptor: | Cyclin-dependent kinase 2, ~{N}-(1-methylpyrazol-4-yl)-5-phenyl-pyrazolo[1,5-a]pyrimidine-7-carboxamide | | Authors: | Watt, J.E, Martin, M.P, Noble, M.E.N. | | Deposit date: | 2022-04-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Build-Couple-Transform: A Paradigm for Lead-like Library Synthesis with Scaffold Diversity.
J.Med.Chem., 65, 2022
|
|
2BYJ
 
 | | Ornithine aminotransferase mutant Y85I | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Markova, M, Peneff, C, Hewlins, M.J.E, Schirmer, T, John, R.A. | | Deposit date: | 2005-08-02 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Determinants of Substrate Specificity in Omega-Aminotransferases.
J.Biol.Chem., 280, 2005
|
|
8A16
 
 | | Human PTPRM domains FN3-4, in spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Caroe, E, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8A1F
 
 | | Human PTPRK N-terminal domains MAM-Ig-FN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hay, I.M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8A17
 
 | | Human PTPRM domains FN3-4, in spacegroup P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase mu, ... | | Authors: | Shamin, M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-05-31 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8DAJ
 
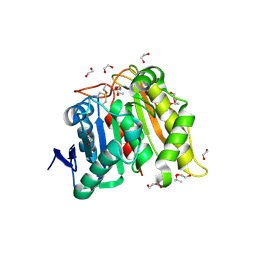 | | Structure and Biochemistry of a Promiscuous Thermophilic Polyhydroxybutyrate Depolymerase from Lihuaxuella thermophilia | | Descriptor: | 1,2-ETHANEDIOL, Esterase, PHB depolymerase family, ... | | Authors: | Thomas, G.M, Quirk, S, Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2022-06-13 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bioplastic degradation by a polyhydroxybutyrate depolymerase from a thermophilic soil bacterium.
Protein Sci., 31, 2022
|
|
8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
8AT1
 
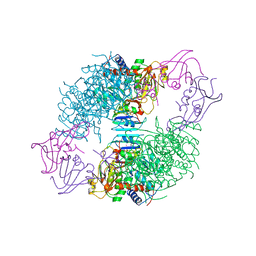 | | CRYSTAL STRUCTURES OF ASPARTATE CARBAMOYLTRANSFERASE LIGATED WITH PHOSPHONOACETAMIDE, MALONATE, AND CTP OR ATP AT 2.8-ANGSTROMS RESOLUTION AND NEUTRAL P*H | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Gouaux, J.E, Stevens, R.C, Lipscomb, W.N. | | Deposit date: | 1989-09-22 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-A resolution and neutral pH.
Biochemistry, 29, 1990
|
|
8ACH
 
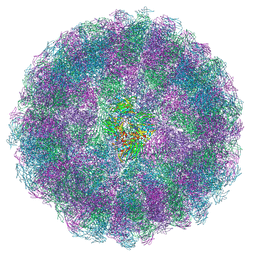 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
