7P3J
 
 | | EED in complex with compound 4 | | Descriptor: | 8-[6-[(dimethylamino)methyl]-2-methyl-pyridin-3-yl]-5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methylamino]-2H-pyrido[3,4-d]pyridazin-1-one, MAGNESIUM ION, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
7P3C
 
 | | EED in complex with compound 4 | | Descriptor: | N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N-[5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methylamino]-[1,2,4]triazolo[4,3-c]pyrimidin-8-yl]benzamide, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
7P3G
 
 | | EED in complex with compound 4 | | Descriptor: | N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-N8-methyl-N8-(1-methylpyrazol-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidine-5,8-diamine, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
6YX9
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) at 2.4 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXD
 
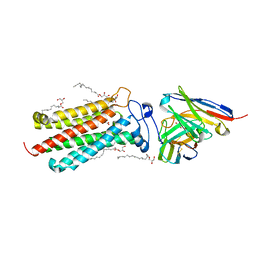 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXG
 
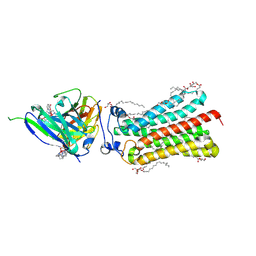 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Tb-XO4 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, OLEIC ACID, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXH
 
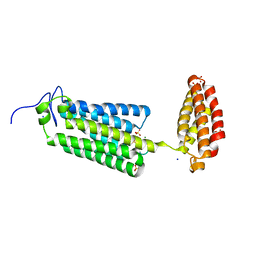 | | Cryogenic human alkaline ceramidase 3 (ACER3) at 2.6 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | Alkaline ceramidase 3, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXF
 
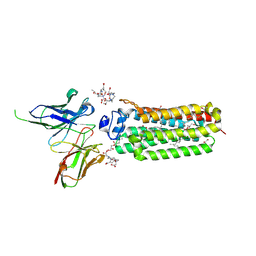 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Adiponectin receptor protein 2, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
7PAG
 
 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
6ZCU
 
 | | syk in complex with 57262_SYKB-AZ13344324-2 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCP
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b | | Descriptor: | 7-[[4-[4-[(dimethylamino)methyl]pyrazol-1-yl]pyrimidin-2-yl]amino]-~{N},3-dimethyl-imidazo[1,5-a]pyridine-1-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 4
To Be Published
|
|
6ZCS
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 3 | | Descriptor: | (3~{R},4~{R})-~{N}4-[1-[2-(1-methylindol-4-yl)-3~{H}-imidazo[4,5-b]pyridin-7-yl]pyrazol-4-yl]oxane-3,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 3
To Be Published
|
|
6ZCX
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 18 | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},1-dimethyl-5-[[4-[3-methyl-4-[[(3~{R})-piperidin-3-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]indazole-3-carboxamide | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 18
To Be Published
|
|
6ZCQ
 
 | | SYK Kinase domain in complex with diamine inhibitor 5 | | Descriptor: | 7-[[4-[4-[[(3~{R},4~{R})-3-azanyloxan-4-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]-~{N},3-dimethyl-imidazo[1,5-a]pyridine-1-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Read, J.A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | SYK Kinase domain in complex with diamine inhibitor 5
To Be Published
|
|
6ZCR
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 7 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 7
To Be Published
|
|
6ZC0
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},~{N}-dimethyl-1-[1-[2-(1-methylindol-4-yl)-3~{H}-imidazo[4,5-b]pyridin-7-yl]pyrazol-4-yl]methanamine | | Authors: | Read, J.A. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCY
 
 | | SYK Kinase domain in complex with diamine inhibitor 16 | | Descriptor: | Tyrosine-protein kinase SYK, ~{N},1-dimethyl-5-[[4-[3-methyl-4-[[(3~{R})-pyrrolidin-3-yl]amino]pyrazol-1-yl]pyrimidin-2-yl]amino]indazole-3-carboxamide | | Authors: | Read, J.A. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 16
To Be Published
|
|
6Z67
 
 | | FtsE structure of Streptococcus pneumoniae in complex with AMPPNP at 2.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, j.A. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
6XP9
 
 | |
7P92
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain up | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P8N
 
 | | TmHydABC- T. maritima hydrogenase with bridge closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P91
 
 | | TmHydABC- T. maritima bifurcating hydrogenase with bridge domain closed | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7P5H
 
 | | TmHydABC- D2 map | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, Fe-hydrogenase, ... | | Authors: | Furlan, C, Chongdar, N, Gupta, P, Lubitz, W, Ogata, H, Blaza, J.N, Birrell, J.A. | | Deposit date: | 2021-07-14 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural insight on the mechanism of an electron-bifurcating [FeFe] hydrogenase.
Elife, 11, 2022
|
|
7OXW
 
 | | CrabP2 mutant R30DK31D | | Descriptor: | ACETATE ION, Cellular retinoic acid-binding protein 2, SULFATE ION | | Authors: | Pastok, M.W, Basle, A, Endicott, J.A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural requirements for the specific binding of CRABP2 to cyclin D3
To Be Published
|
|
6ZSL
 
 | | Crystal structure of the SARS-CoV-2 helicase at 1.94 Angstrom resolution | | Descriptor: | PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Arrowsmith, C.H, von Delft, F, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
