5JGY
 
 | | Crystal structure of maize AKR4C13 in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]oxy}butanoic acid (non-preferred name), Aldose reductase, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of maize AKR4C13 in P21 space group
To Be Published
|
|
6I59
 
 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5XC5
 
 | | Crystal structure of Acanthamoeba polyphaga mimivirus Rab GTPase in complex with GTP | | Descriptor: | ACETATE ION, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ku, B, You, J.A, Kim, S.J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystal structures of two forms of the Acanthamoeba polyphaga mimivirus Rab GTPase
Arch. Virol., 162, 2017
|
|
5XGQ
 
 | |
6I5C
 
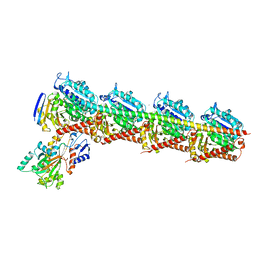 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
5XET
 
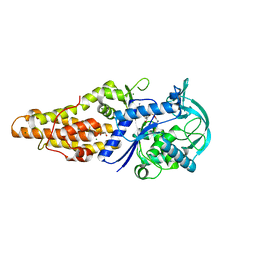 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
5XEQ
 
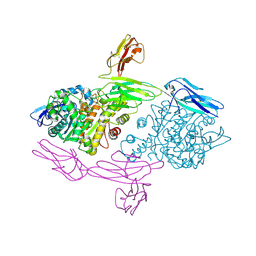 | | Crystal Structure of human MDGA1 and human neuroligin-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, ... | | Authors: | Kim, H.M, Kim, J.A, Kim, D. | | Deposit date: | 2017-04-05 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.136 Å) | | Cite: | Structural Insights into Modulation of Neurexin-Neuroligin Trans-synaptic Adhesion by MDGA1/Neuroligin-2 Complex
Neuron, 94, 2017
|
|
5XC3
 
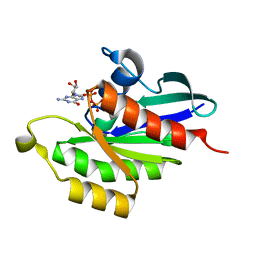 | |
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN0
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
8GHR
 
 | | Structure of human ENPP1 in complex with variable heavy domain VH27.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Carozza, J.A, Wang, H, Solomon, P.E, Wells, J.A, Li, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of VH domains that allosterically inhibit ENPP1.
Nat.Chem.Biol., 20, 2024
|
|
7NZQ
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7O49
 
 | |
7O4A
 
 | |
7O4C
 
 | |
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8SF8
 
 | | Structure of bovine PKA bound to (R)-N-(4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl)-2-amino-4-methylpentanamide | | Descriptor: | N-[4-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]-D-leucinamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Coker, J.A, Arya, T, Goins, C.M, Maw, J.J, Macdonald, J.D, Stauffer, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
J.Med.Chem., 67, 2024
|
|
5O6J
 
 | | Human NMT1 in complex with myristoyl-CoA and inhibitor IMP-1031 | | Descriptor: | 1-[5-[3-fluoranyl-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-1-methyl-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, Glycylpeptide N-tetradecanoyltransferase 1, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2017-06-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
5O7I
 
 | | ERK5 in complex with a pyrrole inhibitor | | Descriptor: | 4-(2-bromanyl-6-fluoranyl-phenyl)carbonyl-~{N}-pyridin-3-yl-1~{H}-pyrrole-2-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J.A, Heptinstall, A, Myers, S. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5O6H
 
 | | Human NMT1 in complex with myristoyl-CoA and inhibitor IMP-917 | | Descriptor: | 1-[5-[4-fluoranyl-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-2~{H}-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2017-06-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
5O48
 
 | | P.vivax NMT with an aminomethylindazole inhibitor bound | | Descriptor: | 1-[5-(4-fluoranyl-2-methyl-phenyl)-1~{H}-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2017-05-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
7USL
 
 | | Integrin alphaM/beta2 ectodomain in complex with adenylate cyclase toxin RTX751 and M1F5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for non-canonical integrin engagement by Bordetella adenylate cyclase toxin.
Cell Rep, 40, 2022
|
|
