4XZV
 
 | | Crystal Structure of SLMO1-TRIAP1 Complex | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, Protein slowmo homolog 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
8TEA
 
 | |
6FFI
 
 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
8SPK
 
 | | Crystal structure of Antarctic PET-degrading enzyme | | Descriptor: | Lipase 1, MALONATE ION | | Authors: | Furtado, A.A, Blazquez-Sanchez, P, Grinen, A, Vargas, J.A, Leonardo, D.A, Sculaccio, S.A, Pereira, H.M, Diez, B, Garratt, R.C, Ramirez-Sarmiento, C.A. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the catalytic activity of an Antarctic PET-degrading enzyme by loop exchange.
Protein Sci., 32, 2023
|
|
4YKQ
 
 | | Heat Shock Protein 90 Bound to CS301 | | Descriptor: | 1-(5-ethyl-2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS301
To Be Published
|
|
6FZS
 
 | | Crystal structure of Smad5-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
6GU2
 
 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GI4
 
 | |
6GUB
 
 | | CDK2/CyclinA in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GL8
 
 | | Crystal structure of Bcl-2 in complex with the novel orally active inhibitor S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Casara, P, Davidson, J, Claperon, A, Le Toumelin-Braizat, G, Vogler, M, Bruno, A, Chanrion, M, Lysiak-Auvity, G, Le Diguarher, T, Starck, J.B, Chen, I, Whitehead, N, Graham, C, Matassova, N, Dokurno, P, Pedder, C, Wang, Y, Qiu, S, Girard, A.M, Schneider, E, Grave, F, Studeny, A, Guasconi, G, Rocchetti, F, Maiga, S, Henlin, J.M, Colland, F, Kraus-Berthier, L, Le Gouill, S, Dyer, M.J.S, Hubbard, R, Wood, M, Amiot, M, Cohen, G.M, Hickman, J.A, Morris, E, Murray, J, Geneste, O. | | Deposit date: | 2018-05-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth.
Oncotarget, 9, 2018
|
|
6GU3
 
 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GXX
 
 | | Fab fragment of an antibody selective for alpha-1-antitrypsin in the native conformation | | Descriptor: | FAB 1D9 heavy chain, FAB 1D9 light chain, MAGNESIUM ION | | Authors: | Elliston, E.L.K, Miranda, E, Perez, J, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterisation of a monoclonal antibody conformationally-selective for native alpha-1-antitrypsin
To Be Published
|
|
6GY4
 
 | | Crystal structure of the N-terminal KH domain of human BICC1 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, CHLORIDE ION, Protein bicaudal C homolog 1, ... | | Authors: | Newman, J.A, Katis, V.L, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of the N-terminal KH domain of human BICC1
To Be Published
|
|
8IJ6
 
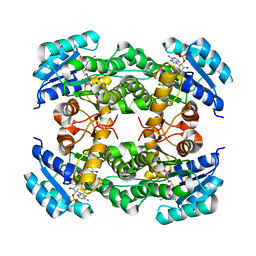 | | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
8IJ8
 
 | |
8IJ7
 
 | |
6GUF
 
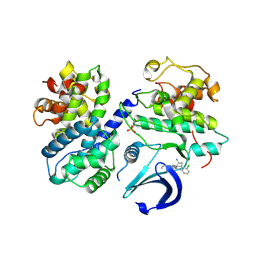 | | CDK2/CyclinA in complex with CGP74514A | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GVK
 
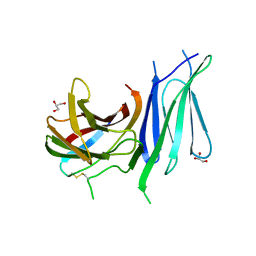 | | Second pair of Fibronectin type III domains of integrin beta4 (T1663R mutant) bound to the bullous pemphigoid antigen BP230 (BPAG1e) | | Descriptor: | Dystonin, GLYCEROL, Integrin beta-4 | | Authors: | Manso, J.A, Gomez-Hernandez, M, Alonso-Garcia, N, de Pereda, J.M. | | Deposit date: | 2018-06-21 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integrin alpha 6 beta 4 Recognition of a Linear Motif of Bullous Pemphigoid Antigen BP230 Controls Its Recruitment to Hemidesmosomes.
Structure, 27, 2019
|
|
8G2E
 
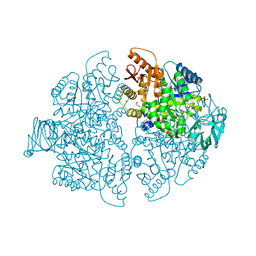 | | PKM2 bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-aminophenyl)methyl]-5-methyl-7-[methyl(oxidanyl)-$l^{3}-sulfanyl]pyridazino[4,5-b]indol-4-one, MAGNESIUM ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Development of Novel Small-Molecule Activators of Pyruvate Kinase Muscle Isozyme 2, PKM2, to Reduce Photoreceptor Apoptosis.
Pharmaceuticals, 16, 2023
|
|
6GU7
 
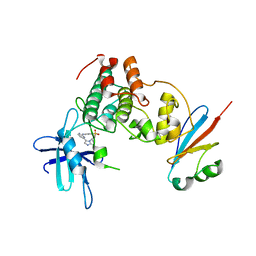 | | CDK1/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GNF
 
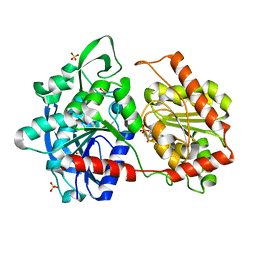 | | Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase, ... | | Authors: | Cuesta-Seijo, J.A, Nielsen, M.M, Palcic, M.M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of theCatalyticDomain ofArabidopsis thalianaStarch Synthase IV, of Granule Bound Starch Synthase From CLg1 and of Granule Bound Starch Synthase I ofCyanophora paradoxaIllustrate Substrate Recognition in Starch Synthases.
Front Plant Sci, 9, 2018
|
|
8IJG
 
 | | Crystal structure of alcohol dehydrogenase M5 from Burkholderia gladioli with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative short-chain dehydrogenases/reductase family protein | | Authors: | Han, X, Mei, Z.L, Liu, W.D, Sun, Z.T, Ma, J.A. | | Deposit date: | 2023-02-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of alcohol dehydrogenase from Burkholderia gladioli with NADP
To Be Published
|
|
6GZV
 
 | | Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Geraets, J.A, Flatt, J.W, Domanska, A, Butcher, S.J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses.
Plos Biol., 17, 2019
|
|
6H5O
 
 | |
6H3E
 
 | | Receptor-bound Ghrelin conformation | | Descriptor: | Appetite-regulating hormone, octan-1-amine | | Authors: | Ferre, G, Damian, M, M'Kadmi, C, Saurel, O, Czaplicki, G, Demange, P, Marie, J, Fehrentz, J.A, Baneres, J.L, Milon, A. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
