7SO4
 
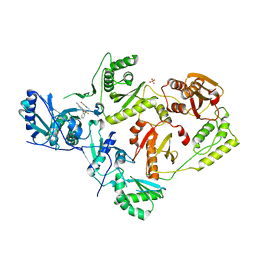 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | 分子名称: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | 著者 | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
5D7D
 
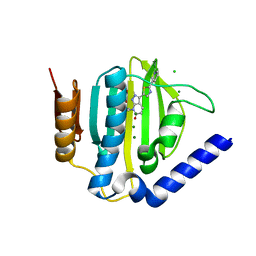 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5D0J
 
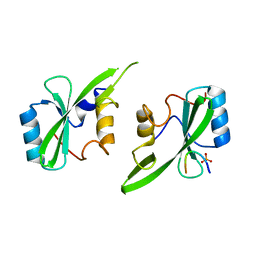 | | Grb7 SH2 with inhibitor peptide | | 分子名称: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | 著者 | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | 登録日 | 2015-08-03 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
7T9H
 
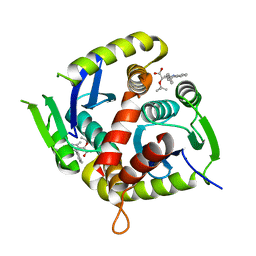 | | HIV Integrase in complex with Compound-15 | | 分子名称: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | 著者 | Khan, J.A, Lewis, H, Kish, K. | | 登録日 | 2021-12-19 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | 著者 | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | 登録日 | 2022-01-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
4R3P
 
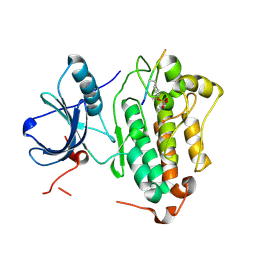 | | Crystal structures of EGFR in complex with Mig6 | | 分子名称: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | 著者 | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | 登録日 | 2014-08-17 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.905 Å) | | 主引用文献 | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7T9O
 
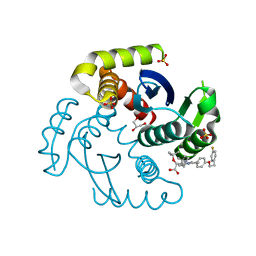 | | HIV Integrase in complex with Compound-25 | | 分子名称: | (2S)-tert-butoxy[4-(4,4-dimethylpiperidin-1-yl)-5-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,6-dimethylpyridin-3-yl]acetic acid, GLYCEROL, Integrase, ... | | 著者 | Khan, J.A, Lewis, H, Kish, K. | | 登録日 | 2021-12-19 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7THB
 
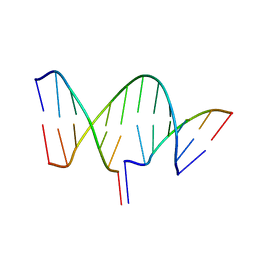 | | Crystal structure of an RNA-5'/DNA-3' strand exchange junction | | 分子名称: | DNA (5'-D(*GP*AP*TP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*TP*AP*AP*GP*CP*AP*GP*CP*AP*TP*C)-3'), RNA (5'-R(*AP*GP*CP*UP*UP*AP*C)-3') | | 著者 | Cofsky, J.C, Knott, G.J, Gee, C.L, Doudna, J.A. | | 登録日 | 2022-01-10 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of an RNA/DNA strand exchange junction.
Plos One, 17, 2022
|
|
5D7C
 
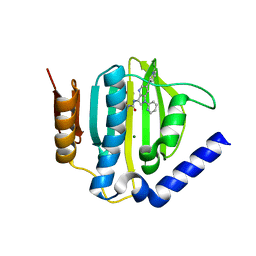 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D7R
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-14 | | 公開日 | 2015-11-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
7T1T
 
 | | JAK2 JH2 IN COMPLEX WITH JAK292 | | 分子名称: | (2S)-2-[({4-[(2-amino-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}carbamoyl)amino]-4-phenylbutanoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | 著者 | Ippolito, J.A, Henry, S, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | 登録日 | 2021-12-02 | | 公開日 | 2022-05-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Conversion of a False Virtual Screen Hit into Selective JAK2 JH2 Domain Binders Using Convergent Design Strategies
Acs Med.Chem.Lett., 13, 2022
|
|
4R6K
 
 | | Crystal structure of ABC transporter substrate-binding protein YesO from Bacillus subtilis, TARGET EFI-510761, an open conformation | | 分子名称: | SODIUM ION, SOLUTE-BINDING PROTEIN | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-25 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of transporter Yeso from Bacillus subtilis, Target Efi-510761
To be Published
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | 分子名称: | Alpha-synuclein | | 著者 | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | 登録日 | 2014-10-06 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | 主引用文献 | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
7B9B
 
 | | Crystal structure of human 5' exonuclease Appollo APO form | | 分子名称: | 5' exonuclease Apollo, NICKEL (II) ION | | 著者 | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-12-14 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
7T0P
 
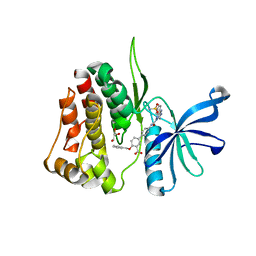 | | JAK2 JH2 IN COMPLEX WITH JAK315 | | 分子名称: | 4'-{[5-amino-3-(4-sulfamoylanilino)-1H-1,2,4-triazole-1-carbonyl]amino}-4-(benzyloxy)[1,1'-biphenyl]-3-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | 著者 | Ippolito, J.A, Liosi, M.-E, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | 登録日 | 2021-11-30 | | 公開日 | 2022-06-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Insights on JAK2 Modulation by Potent, Selective, and Cell-Permeable Pseudokinase-Domain Ligands.
J.Med.Chem., 65, 2022
|
|
8ACT
 
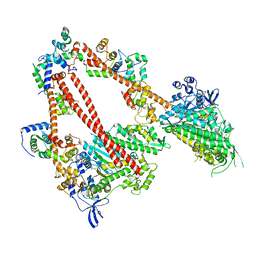 | | structure of the human beta-cardiac myosin folded-back off state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | 著者 | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | 登録日 | 2022-07-06 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
4RKF
 
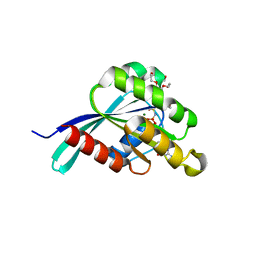 | | Drosophila melanogaster Rab3 bound to GMPPNP | | 分子名称: | MAGNESIUM ION, PENTAETHYLENE GLYCOL, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Lardong, J.A, Driller, J.H, Depner, H, Weise, C, Petzoldt, A, Wahl, M.C, Sigrist, S.J, Loll, B. | | 登録日 | 2014-10-13 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4RXU
 
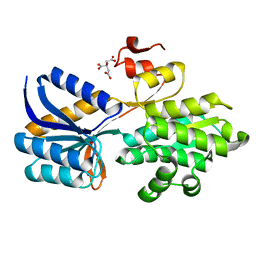 | | Crystal structure of carbohydrate transporter solute binding protein CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158, in complex with D-glucose | | 分子名称: | CITRIC ACID, Periplasmic sugar-binding protein, beta-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-12-11 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of sugar transporter CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158
To be Published
|
|
4RY1
 
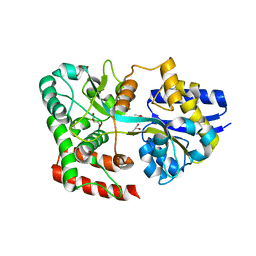 | | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum SCRI1043, Target EFI-510858 | | 分子名称: | ACETATE ION, GLYCEROL, Periplasmic solute binding protein | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-12-12 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of periplasmic solute binding protein ECA2210 from Pectobacterium atrosepticum, Target EFI-510858
To be Published
|
|
7TVA
 
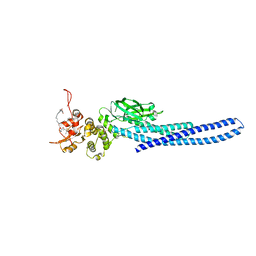 | | Stat5a Core in complex with AK2292 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.835 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7SZU
 
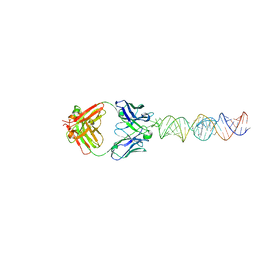 | |
7TVB
 
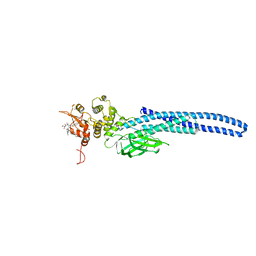 | | Stat5A Core in Complex with AK305 | | 分子名称: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7SVQ
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea in complex with NAD+ | | 分子名称: | L-galactose dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | 登録日 | 2021-11-19 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
8AGS
 
 | | Cyclohexane epoxide soak of epoxide hydrolase from metagenomic source ch65 resulting in halogenated compound in the active site | | 分子名称: | 1,2-ETHANEDIOL, 2-CHLOROPHENOL, Alpha/beta epoxide hydrolase, ... | | 著者 | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | 登録日 | 2022-07-20 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8AGN
 
 | | Cyclohexane epoxide low pH soak of epoxide hydrolase from metagenomic source ch65 | | 分子名称: | (1R,6S)-7-oxabicyclo[4.1.0]heptane, 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, ... | | 著者 | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | 登録日 | 2022-07-20 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.957 Å) | | 主引用文献 | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
