7O4C
 
 | |
1Z4U
 
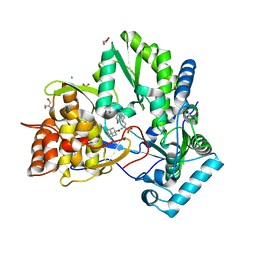 | | hepatitis C virus NS5B RNA-dependent RNA polymerase complex with inhibitor PHA-00799585 | | Descriptor: | (2Z)-2-[(1-ADAMANTYLCARBONYL)AMINO]-3-[4-(2-BROMOPHENOXY)PHENYL]PROP-2-ENOIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pfefferkorn, J.A, Greene, M, Nugent, R, Gross, R.J, Mitchell, M.A, Finzel, B.C, Harris, M.S, Wells, P.A, Shelly, J.A, Anstadt, R. | | Deposit date: | 2005-03-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8SH5
 
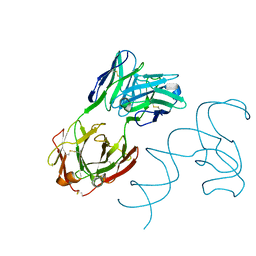 | | Crystal structure of 3'cap-independent translation enhancers (CITE) from Pea enation mosaic virus RNA 2 (PEMV2) with Fab BL3-6K170A | | Descriptor: | Fab BL3-6K170A heavy chain, Fab BL3-6K170A light chain, RNA (88-MER) | | Authors: | Lewicka, A, Roman, C, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a cap-independent translation enhancer RNA.
Nucleic Acids Res., 51, 2023
|
|
7NUZ
 
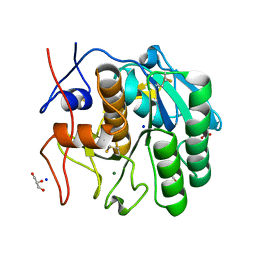 | | Proteinase K structure at atomic resolution from crystals grown in agarose gel | | Descriptor: | GLYCEROL, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Gavira, J.A, Artusio, F, Castellvi, A, Pisano, R. | | Deposit date: | 2021-03-15 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Tuning Transport Phenomena in Agarose Gels for the Control of Protein Nucleation Density and Crystal Form
Crystals, 2021
|
|
7NUY
 
 | |
7O4B
 
 | |
7NZZ
 
 | |
7NWU
 
 | | Co-crystal structure of UPF3B-RRM-NOPS-L with UPF2-MIF4GIII | | Descriptor: | PENTAETHYLENE GLYCOL, Regulator of nonsense transcripts 2, Regulator of nonsense transcripts 3B, ... | | Authors: | Powers, K.T, Bufton, J.C, Szeto, J.A, Schaffitzel, C. | | Deposit date: | 2021-03-17 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of nonsense-mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation.
Nucleic Acids Res., 50, 2022
|
|
7NTM
 
 | |
7OIO
 
 | |
8FE5
 
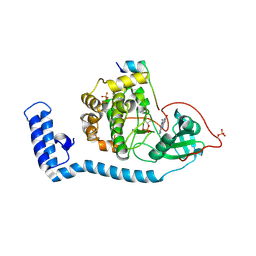 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
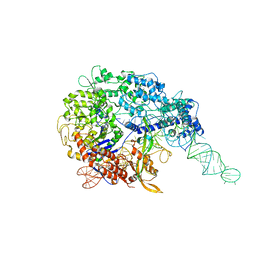
6MCC
 
 | |
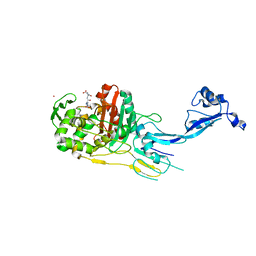
7OK9
 
 | |
2ACL
 
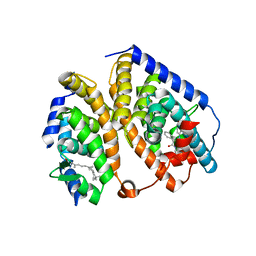 | | Liver X-Receptor alpha Ligand Binding Domain with SB313987 | | Descriptor: | 1-BENZYL-3-(4-METHOXYPHENYLAMINO)-4-PHENYLPYRROLE-2,5-DIONE, Oxysterols receptor LXR-alpha, RETINOIC ACID, ... | | Authors: | Jaye, M.C, Krawiec, J.A, Campobasso, N, Smallwood, A, Qiu, C, Lu, Q, Kerrigan, J.J. | | Deposit date: | 2005-07-19 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of substituted maleimides as liver x receptor agonists and determination of a ligand-bound crystal structure.
J.Med.Chem., 48, 2005
|
|
7OOL
 
 | | Crystal structure of a Candidatus photodesmus katoptron thioredoxin chimera containing an ancestral loop | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Ibarra-Molero, B, Gamiz-Arco, G, Risso, V, Sanchez-Ruiz, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining Ancestral Reconstruction with Folding-Landscape Simulations to Engineer Heterologous Protein Expression.
J.Mol.Biol., 433, 2021
|
|
7P3J
 
 | | EED in complex with compound 4 | | Descriptor: | 8-[6-[(dimethylamino)methyl]-2-methyl-pyridin-3-yl]-5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methylamino]-2H-pyrido[3,4-d]pyridazin-1-one, MAGNESIUM ION, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
7P3C
 
 | | EED in complex with compound 4 | | Descriptor: | N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N-[5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methylamino]-[1,2,4]triazolo[4,3-c]pyrimidin-8-yl]benzamide, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
7P3G
 
 | | EED in complex with compound 4 | | Descriptor: | N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, N5-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-N8-methyl-N8-(1-methylpyrazol-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidine-5,8-diamine, Polycomb protein EED | | Authors: | Read, J.A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Diverse, Potent, and Efficacious Inhibitors That Target the EED Subunit of the Polycomb Repressive Complex 2 Methyltransferase.
J.Med.Chem., 64, 2021
|
|
6M9I
 
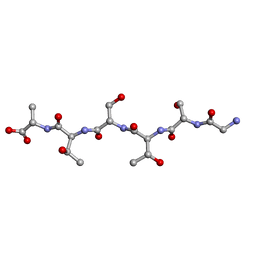 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
7PAG
 
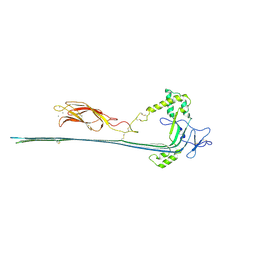 | | The pore conformation of lymphocyte perforin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Perforin-1 | | Authors: | Ivanova, M.E, Lukoyanova, N, Malhotra, S, Topf, M, Trapani, J.A, Voskoboinik, I, Saibil, H.R. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The pore conformation of lymphocyte perforin.
Sci Adv, 8, 2022
|
|
6MCD
 
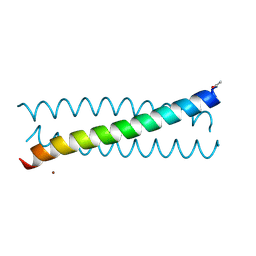 | | Crystal Structure of tris-thiolate Pb(II) complex with adjacent water in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | LEAD (II) ION, Pb(II)(GRAND Coil Ser L12CL16A)-, ZINC ION | | Authors: | Tolbert, A.E, Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-31 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteromeric three-stranded coiled coils designed using a Pb(II)(Cys)3template mediated strategy.
Nat.Chem., 12, 2020
|
|
6MG9
 
 | |
6M40
 
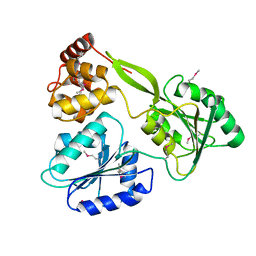 | | Crystal structure of the NS3-like helicase from Alongshan virus | | Descriptor: | NS3-like protein | | Authors: | Gao, X.P, Zhu, K.X, Chen, P, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2020-03-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of the NS3-like helicase from Alongshan virus.
Iucrj, 7, 2020
|
|
6MCB
 
 | |
6M7M
 
 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
